Technical manual of the application HD4DP v2
Technical manual of the application HD4DP v2The content of this page is only available in EN. Select the EN language button to read it.
Technical user roles in HD4DP v2
Technical user roles in HD4DP v2IT administrator: An IT administrator has the highest level of all roles and permissions and can:
- log in using Active Director;
- grant access to Local Study Lead, Local Study Associate and Local Study Support;
- select and access all projects;
- create, find, update, delete, send (to healthdata.be, MyCareNet and other destinations) and correct a record using the form.io component;
- create, update, send and correct a record using the API data collection;
- create, update, send and correct a record using CSV upload;
- create and send a MyCareNet record using MyCareNet XML;
- view all records for all projects;
- harvest all records for all projects from the local DWH using the PostgreSQL database.
HD4DP v2 Installation
HD4DP v2 InstallationHD4DP v2 Local is an application installed on the infrastructure of the Health Care Organisation participating in research projects facilitated by healthdata.be.
The installation of HD4DP v2 Local is executed by the DevOps team of healthdata.be.
Server Installation and Configuration
Installing and configuring the server requires the following actions:
- install and configure the server as described in the article "HD4DP 2.0 Infrastructure instructions";
- complete and return to support.healthdata@sciensano.be the server configuration file "HD4DP v2 Infrastructure Sheet".
The HD4DP v2 application is more modular and will support scaling up to meet the requirements of the various data collection projects we facilitate. It will offer several micro-services that will run concurrently on the same machine.
The server should therefore require more resources than the one currently hosting the HD4DP 1.0 application. Furthermore, the resources allocated should be increased. It is therefore on the one hand imperative to use virtualization for the creation of the machine. On the other hand. It is also imperative to store files and make regular backups to a file server.
Below we take up our three categories of organizations sending data to healthdata.be and the resources we recommend allocating to their virtual machine:
- “Small”: Small data provider.
- “Medium”: Medium data provider;
- “Large”: Big data provider.
Finally, we also offer the possibility for each hospital to have an integration server and a production server. Healthdata.be will deploy the new release of the application on the integration server. This will allow you to accept or decline the promotion of a new release of the HD4DP 2.0 application to the production server. This option is highly recommended, but not mandatory.
Therefore, could you answer the question: Do you want to first deploy HD4DP on an integration server? Yes/No. If Yes, Could you provide a server whose label used for specifications is ‘Small’ (following the instructions in section 1 of this mail), that is:
- Processors number: 1
- Physical cores/Processor : 8
- RAM memory : 16 Go
- Disk space: 100 Go
- Network Station Mount with Space for Backups
- Operating System: Linux Ubuntu v18.04
- Virtualization
Server installation timing
In order to establish the deployment schedule for the HD4DP 2.0 application within your organization, we would like to know when the server could be installed and configured. To this end, could you give us the 2 dates relating to the installation of the server:
- Starting date ;
- Finalization date.
Based on these dates, an employee of healthdata.be will regularly monitor the operations linked to the installation of the server.
For any request for information on installing the HD4DP 2.0 server, please send an email to hd-architecture-20@sciensano.be.
HD4DP v2 Infrastructure instructions
HD4DP v2 Infrastructure instructionsIntroduction
This document is written for IT staff / system engineers of data providers and therefore assumes technical knowledge. It acts as a guide through the on-boarding process of HD4DP v2 and covers installation of the server, user configuration, network configuration and remote access.
The order of steps in this document should be respected during execution.
Overview
HD4DP v2 consists of a modular application stack, which allows healthdata.be to seamlessly upgrade individual elements.
An HD4DP v2 deployment comprises of following components:
- Form.io component
- MongoDB
- PostgreSQL
- Nextgen Connect
As it is the case in HD4DP 1.0, an Encryption Module with a connection to the eHealthBox is still required for HD4DP v2 and must be provided by the data provider.
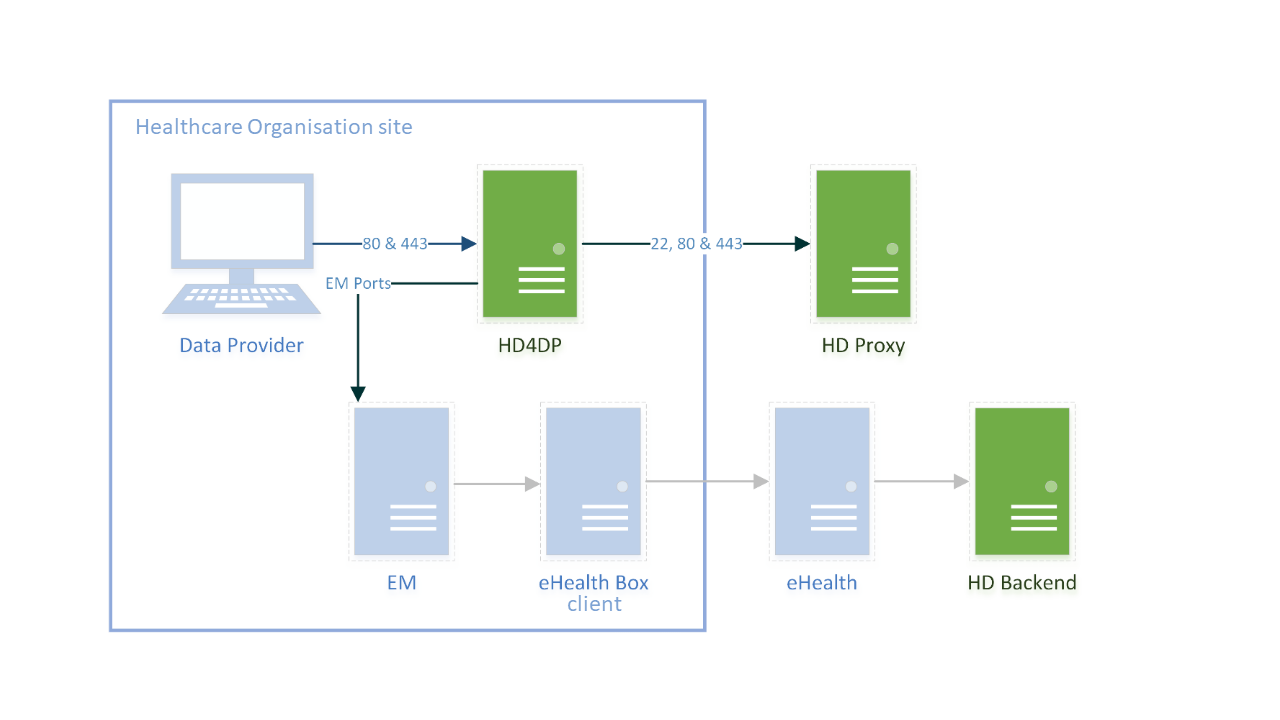
Network configuration
IP
The HD4DP server needs to be accessible via domain names in DNS, and must have a static IP in your private network.
DNS
The application stack of HD4DP v2 requires four domain names pointing to the IP of the locally installed HD4DP v2 server. Use the following names in your DNS:
- nextgenconnect.hd4dp.<yourdomain.be>
- hd4dp.<yourdomain.be>
- metabase.hd4dp.<yourdomain.be>
- admin.hd4dp.<yourdomain.be>
Firewall
The following connections should be possible in the firewall flow:
- To and from (a) machine(s) in your IT department on port 22 for initial configuration and local support.
- To and from the Encryption Module server. The protocol and ports depend on your local EM implementation. Contact your EM vendor if more information is necessary.
- Reachable by your staff who uses HD4DP, on ports 80 and 443 for HTTP(s) traffic.
- To and from the LDAP server (this is not mandatory if you are not using LDAP to authenticate) (port 389 by default)
The healthdata.be proxy server is used as a gateway to the internet for the security of HD4DP servers. The configuration of this proxy server will be provided to you by healthdata.be at a later date.
Server installation
To install the application stack of HD4DP v2, healthdata.be requires a fresh installed operating system, specifically Ubuntu Server 18.04 LTS.
Please use these instructions even if you have previous experience with installing this operating system, as its configuration is specific for healthdata.be.
These instructions assume that the network configuration described in the previous section is completed.
Instructions
HD4DP v2 requires a (virtual) machine running Ubuntu Server 18.04 LTS.
We assume knowledge of loading a .iso file onto a (virtual) machine. Healthdata.be can’t provide instructions for this, as the environment of your center is unknown. Should you have any trouble, however, please contact Healthdata.be support so that we can help out.
Please find the installation steps below.
Installation steps
- Download the .iso file from the link below.
Download Ubuntu Server 18.04 LTS - Create a new (virtual) machine with Linux Ubuntu 64 bit as the OS family
- When prompted, select the .iso file downloaded in step 1.
- After some time, you will be prompted to select a system language. Select English.
- “Keyboard configuration”
Select your preferred keyboard layout and press enter - “Network Connections”
Highlight the network interface and press enter. Navigate as follows:
Edit IPv4 -> Manual -> enter the network details -> save -> Done - Proxy IP -> Leave default/empty.
- “Configure Ubuntu Archive Mirror” -> leave default
- “File system Setup” -> Use An Entire Disk
- Proceed until “Confirm destructive action” -> press continue. The installation process starts, this can take several minutes.
- In the meantime, create the user for Healthdata.
username = healthdata,
Password = choose a secure password and communicate it to healthdata.be. - Mark “Install OpenSSH server”. This will be used for remote access. “Import SSH Identity” -> no -> done
- “Featured Server Snaps” -> Select nothing and press Done.
- Wait until installation is finished.
Configuration steps
Connecting to the server
Log into the machine with the healthdata.be user created in the previous section.
Instructions (from a Windows machine):
- Install the tool Putty and open the application.
- On the configuration screen, enter the following (replace cursive text with the appropriate values)
- Host Name: healthdata@server_private_ip
- Port: 22
- Connection type: SSH
- Click Open. Enter the password (you will not see text as you type, you can paste into putty by right-clicking in the terminal).
- You should now be logged in and see a prompt “healthdata@server_name:~$”
Administrator account for internal use
An administrator account for internal use can be created on the HD4DP v2 server.
The configuration of remote access (described below) should not happen on this account, but on the Healthdata.be account.
The internal account can later be used to install and configure OS monitoring software and antivirus software by the internal IT team. For more information, see the section on Antivirus and Monitoring.
(Text with a gray background should be entered as a command in the terminal of the server)
Create the user:
sudo adduser <username>
Add the user to the sudo group
sudo usermod -aG sudo <username>
Installation and configuration of the software stack
Healthdata.be support will instruct you when to execute the next step, which is to enable remote access so that Healthdata.be can execute the software installation and configuration.
Backups
The configuration of the HD4DP v2 server is administered by healthdata.be and does not require backups.
HD4DP v2 regularly dumps its databases automatically to the /backup directory on the server. A network storage should be mounted at this location.
Please fill out the infrastructure sheet with the required credentials, domain name/url, protocol… to connect to the network drive. The connection will then be configured by healthdata.be.
Patching and Updates
Healthdata.be configures HD4DP v2 servers to automatically receive recommended security updates. The choice for Ubuntu 18.04 is motivated by the long-term support for this version. Security flaws are rare in this distribution, and security updates are quick and often don’t require a system reboot.
If the IT department of your organization prefers to manage patches, this is possible but not encouraged. Please use the account for internal use created in Section 3 for this purpose.
Antivirus and Monitoring
Most data providers will want to manage their own antivirus and OS monitoring on all machines in their network. Installation of such software on the HD4DP v2 server is allowed, but healthdata.be should be informed about all extra software installed on the server. Additionally, healthdata.be will not provide support for the installation of this software.
Contact information
HD4DP v2 Infrastructure sheet
HD4DP v2 Infrastructure sheetThe HD4DP v2 Infrastructure Sheet contains information that healthdata.be needs in order to start the insallation of the HD4DP 2.0 Software at your organization.
Below you can find the description of the necessary information:
SERVER CONNECTION
Healthdata.be performs its installation and support tasks remotely (using VPN or remote port forwarding via SSH). Please provide the required credentials.
- Type of connection (VPN / Remote port forwarding via SSH)
- Link (IF VPN)
- Username, token, other (if VPN)
- Password (if VPN)³
- Public SSH Key (if remote port forwarding)
³ For security reasons, we advise to communicate passwords to us either by phone, or via a link using a secret-sharing service such as onetimesecret.com.
SERVER MACHINE
- Server Name
- Internal IP-Address
- Ram (in GB)
- CPU (number of CPU's and number of cores)
- Disk space (in GB)
- Username: Healthdata
- Password ³
³ For security reasons, we advise to communicate passwords to us either by phone, or via a link using a secret-sharing service such as onetimesecret.com.
ATTACHED DRIVE FOR BACKUPS
HD4DP 2.0 regularly performs data dumps for backup purposes. Please provide connection information to a network share volume.
- Link / IP address
- Path
- Username
- Password ³
³ For security reasons, we advise to communicate passwords to us either by phone, or via a link using a secret-sharing service such as onetimesecret.com.
USER MANAGEMENT
HD4DP can either connect to a LDAP server or use its own application database for performing user authentication and management. Please check the user management mechanism you want to use.
- LDAP user management : Yes / No
- Application user management : Yes / No
LDAP configuration (Optional)
If you chose ‘LDAP user management’ as user management mechanism, please provide the following information that allows us to connect to your LDAP system.
- Connection URL
- Username
- Password³
³ For security reasons, we advise to communicate passwords to us either by phone, or via a link using a secret-sharing service such as onetimesecret.com.
SOFTWARE CONFIGURATION
Encryption Module interface
HD4DP communicates with the Encryption Module (EM) either using the file system interface or by calling a REST web service. Please choose which interface HD4DP should use for its communication with the Encryption Module.
Note: if the encryption module is not yet purchased (or developed), HD4DP can already be installed; the EM can then be configured in HD4DP once it is available. Please note that HD4DP 1.x and HD4DP 2.0 cannot use the same EM.
- REST web service
- File system
REST web service interface
If you chose to communicate with the Encryption Module using a REST interface, please provide the web service URLs that should be used by HD4DP for its communication with EM.
- "Outgoing flow URL: Example: http://host:8080/encryptionmodule/send"
- "Incoming flow URL : Example: http://host:8080/encryptionmodule/receive"
File system interface
- "Incoming directory: Directory where HD4DP checks for incoming files"
- "Incoming directory: Directory where HD4DP writes outgoing files"
- "Incoming directory: Directory to which HD4DP moves successfully processed files"
- "Incoming directory: Directory to which HD4DP moves unsuccessfully processed files"
Requirements for the HD4DP installation
Requirements for the HD4DP installationThis documentation details the necessary adaptations to be performed in order to allow the necessary technical accesses and smooth operation of the different healthdata.be platforms and interfaces.
The file is available for download below.
HD4DP v2 S2S API
HD4DP v2 S2S APIIntroduction
The HD4DP v2 S2S API is a unified Application Programming Interface (API) that will allow participating Healthcare Organizations (HCO) to submit DCDs data to HD4DP2.0 fully automated.
In the manual of the application HD4DP v2 we provide detailed information about the S2S API. Please read this documentation before its project specific use.
- End-to-End process to submit DCD registrations
- API Endpoint for supporting the DCD submit process
- MDM Field description - DB Model
- Swagger API
- Postman collection
Important note:
For code fields (fieldType = 'CODE') the id of the codeListValue item must be sent, not the code value or the label. In future releases it will be made possible to also send the code value.
Training
Below you can review the S2S API training organized by healthdata.be:
The study project [ ] consists of 1 DCD:
- [ ]
On the following pages we explain how to submit [ ] using the HD4DP v2 S2S API.
This documentation is being updated regularly. We try to provide as correct, complete and clear as possible information on these pages. Nevertheless, if you see anything in the documentation that is not correct, does not match your experience or requires further clarification, please create a request (type : request for information) via our portal (https://sciensano.service-now.com/sp) or send us an e-mail via support.healthdata@sciensano.be to report this documentation issue. Please, do not forget to mention the URL or web address of the page with the documentation issue. We will then adjust the documentation as soon as possible. Thank you!S2S API for IQEDFOOT
S2S API for IQEDFOOTDocumentation for System 2 System API on Architecture 2.0
Description of the service
API is the acronym for Application Programming Interface, which is a software intermediary that allows two applications to talk to each other.
In this case, the API is used to have the system of the Data Providers communicate with the system of HealthData.
The S2S API is a unified API that will allow clients (Data Providers) to submit DCDs data to HD4DP2.0 in fully automated way.
End point information (per DCD) + examples
| API | Response | Notes |
|---|---|---|
| /api/organizations | List of organizations. Client must select the right organizationId | Current existing end-point is: /api/installation/organizations We’ll create this new end-point with a different signature re-routing the call to this existing one or we will refactor the existing one to this new signature. |
| /api/dcd/menu/structure? organization-id={organizationId} | List of projects of the given organization, dcds of each project, dcdVersions of each dcd in a JSON format Client can get dcdId and dcdVersionId (optional) which are needed on following API calls. | |
| /api/dcd/payload/definition? dcd-id={dcdId}; <optional>version={version}; <optional>language-id={languageId} | List of all the fields of the form as well as their corresponding data-types that are allowed in the json data structure for the Payload | This field names values are the key properties in the formIO json config form. When we implement this new api end-point, we need to parse the json content in order to get the key properties. Given these field keys, we’ll get each field definition from new API end-points helpers: /api/dcd/field?field-id={fieldId} /api/dcd/codelist?codelist-id={codelistId} These ones are described in the next table. <optional parameter> version={version} : If this parameter is not provided, latest one is assumed <optional parameter> language-id = {languageId} : language id for the code_list example results. If this parameter is not provided, default language will be English. Current permitted values: en: English nl: Dutch fr: French Client must build this json object as the payload data to be sent based on this list of fields, on the last api call |
| /api/dcd/payload/example?dcd-id={dcdId}; <optional>version={version} | Example of payload in JSON format | Providing this API end-point in order to help the Client on the Payload build with an example <optional parameter> version={version} : If this parameter is not provided, latest one is assumed |
| /api/dcd/payload/submit? organization-id={organizationId}; dcd-id={dcdId}; <optional>version={version}; <optional>data-src-type={dataSrcType}; POST Payload | /api/dcd/payload/submit? organization-id={organizationId}; dcd-id={dcdId}; <optional>version={version}; <optional>data-src-type={dataSrcType}; POST Payload | Some implementation tasks is needed in here in order to return the result info (either succeed or failed). Similar like the one in HDConnectProxyRestTemplate.postCsv method, and the CsvExecutionResult object build. <optional parameter> version={version} : If this parameter is not provided, latest one is assumed. <optional parameter> data-src-type={dataSrcType} : permitted values: API CSV If this parameter is not provided, default values is <HD4DP>. |
| api/dcd/submit?organization-id={organizationtId};dcd-id={dcd-id};<optional>dcd-version-id={dcdVersionId};<optional>incl-submit-data={inclSubmitData}; <optional>incl-submit-results={inclSubmitResults}; GET method | List of submitted dcds data and/or their corresponding business keys or validation errors. | <optional parameter> dcd-version-id={dcdVersionId} : If this parameter is not provided, lastest one is assumed <optional parameter> incl-submit-data={inclSubmitData} : If this parameter is not provided, default values is <false> <optional parameter> incl-submit-results={inclSubmitResults} : If this parameter is not provided, default values is <true> |
HOW TO: Upload data using System 2 System
Steps To Upload data
1. IT services of data providers must setup their systems to be able to communicate with HD4DP v2 System 2 System API
A prerequisite to be able to use the Health Data's System 2 System API, is that the IT services of the hospitals must have the following in place before the systems can communicate:
- The endpoint/URL is protected by credentials for which the support services need to be contacted.
- End-to-end API process to submit DCD registrations in a fully automated manner.
- Support for searching submitted DCD registrations.
2. Prepare the JSON file (example file in this section)
To send DCD registrations to Health Data, the file must be in a .json file format.
- At the Data Provider's side, we must foresee a way to extract the JSON file from the electronic patient files and/or other local databases.
- Author group, Author and Coauthor:
- When the Author group, Author and Coauthor has been left out in the json file, the default Author group, Author and Coauthor will be used automatically.
- When the desired Author group, Author and Coauthor are specified in the json file, the following fields TX_AUTHOR_GR, TX_AUTHOR and TX_COAUTHOR must be added to the json file with their values respectively.
Example:
TX_AUTHOR_GR;TX_AUTHOR;TX_COAUTHOR
Test group;test@sciensano.be;test@sciensano.beNote:
The Author group, Author and Coauthor must exist and are well configured at the back-end of the system. TX_AUTHOR_GR can be a string that identifies the Author group to which this Author belongs. Commonly, the first name and last name are used to identify the TX_AUTHOR_GROUP. Be sure to avoid leading and trailing spaces when entering the Author group value.
- Similarly, to submit a draft, the only thing to add is the key STATUS (all upper case) with the value "draft" to the request, as described in Submission of drafts.
- Adding separators to a NISS number:
It is not necessary to necessary to add separators in a NISS number when uploading a file using S2S API. You can fill the NISS number out both with or without seperators. E.g.: 85.04.02-169.32 or 85040216932.
Example:
- Make sure the name of the JSON file has the correct format:
HD_DCD_submjson_HDBPnumber_HDBPabbreviation_versionnumber_versionreleasedate
So for <dcd name> the format would be:
HD_DCD_submjson_<HDBPnumber>_<HDBPabbreviation>_<versionnumber>_<versionreleasedate>
Please find here an example file for <dcd name>:
<paste the content of the json file here>
Disclaimer: The example files above are only provided as a guideline and do not contain real life data.
3. Uploading the JSON File
- The IT department of the Data Provider must provide a manner in their system to send the API requests containing DCD registrations in a JSON file format, the correct end-points must be addressed.
- In this case the end-point to upload the json file will be:
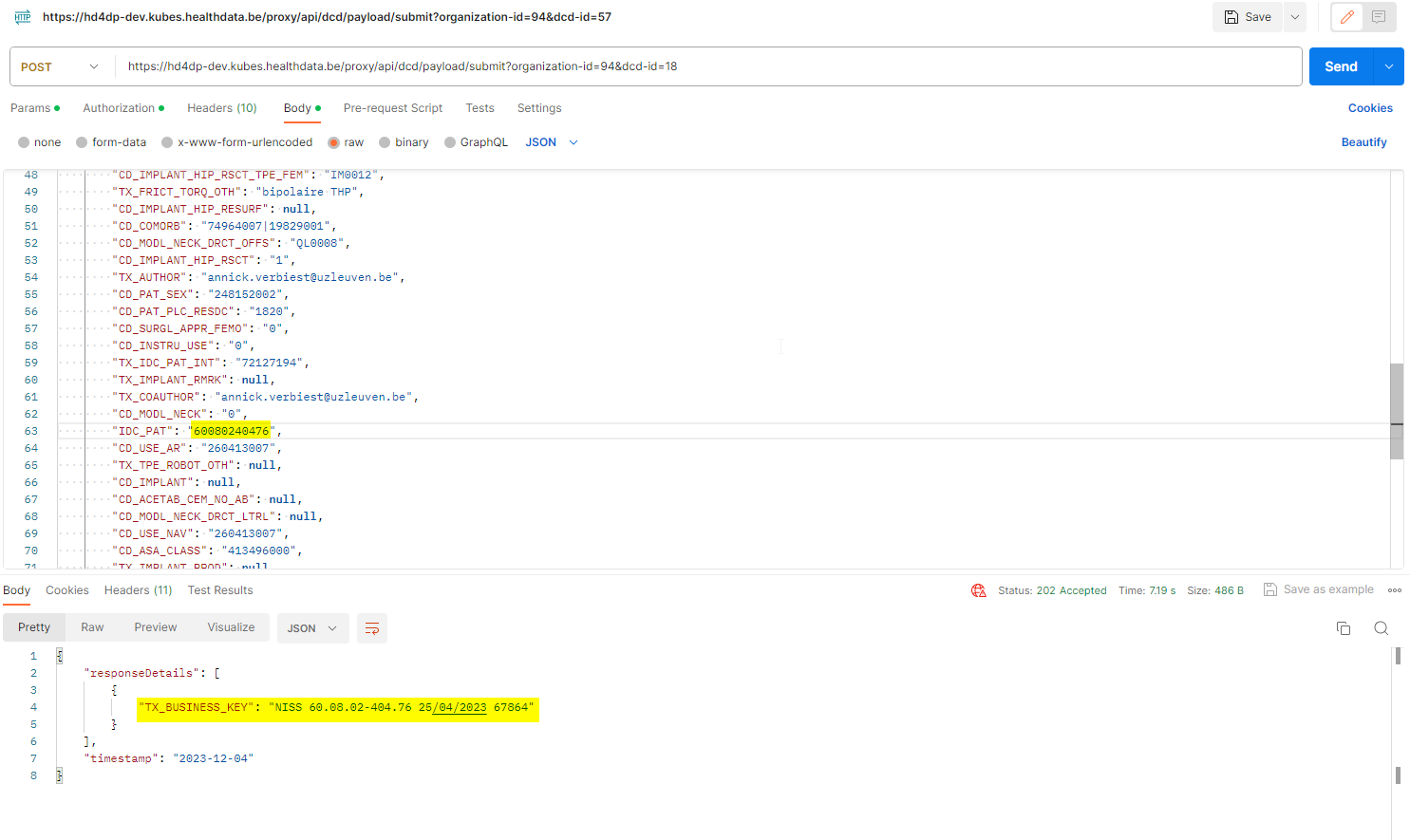
4. Validate the JSON Upload
4.1 Validation of the S2S API Upload via the response:
Verify in the same way the request was sent, that the returned response is containing a valid Business key.
If a valid Business key has been provided, the registration upload via System 2 System API was succesful.
4.2 Validation of the System 2 System API via HD4DP 2.0:
Step 1: Open the web application HD4DP 2.0.
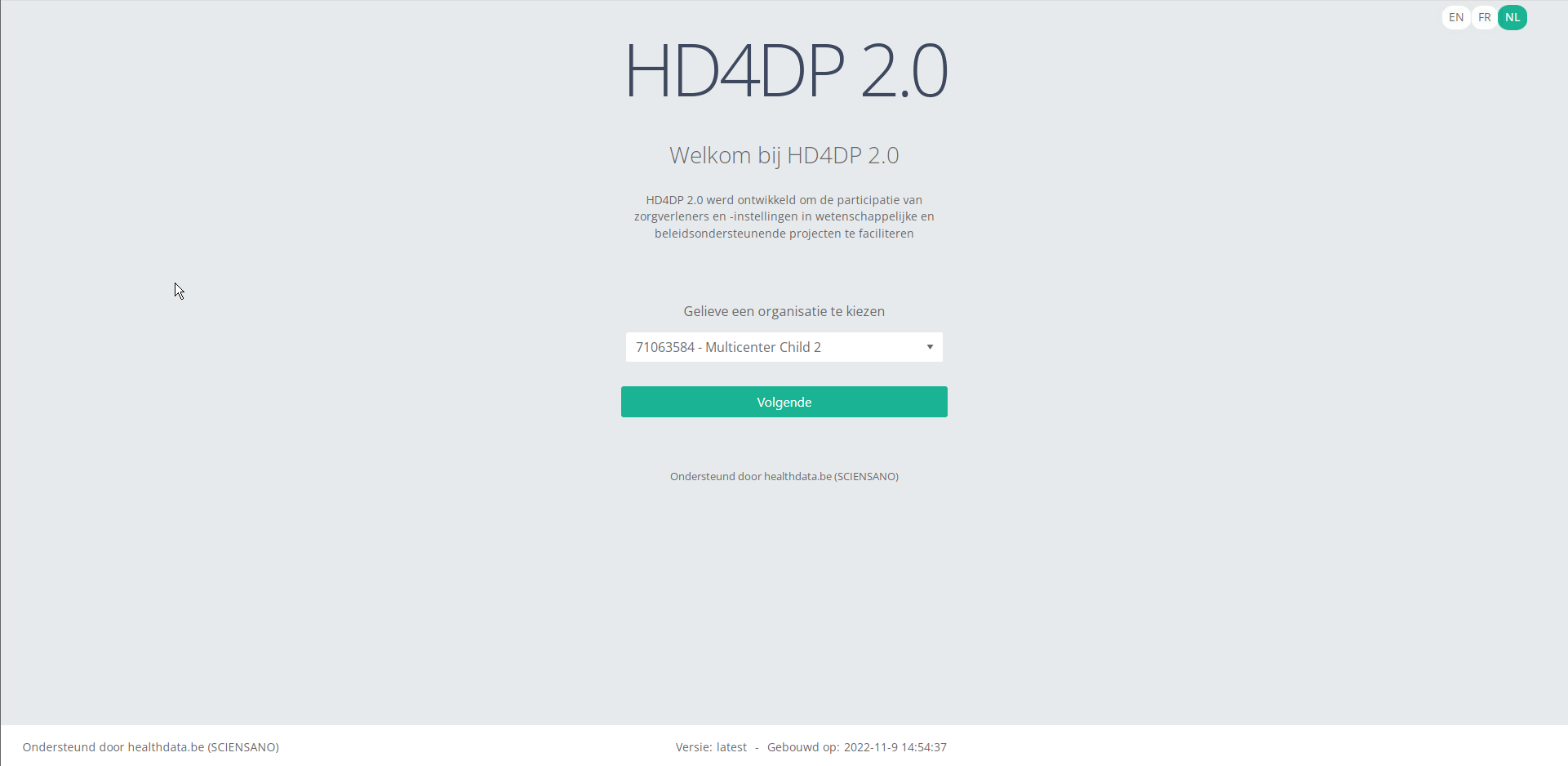
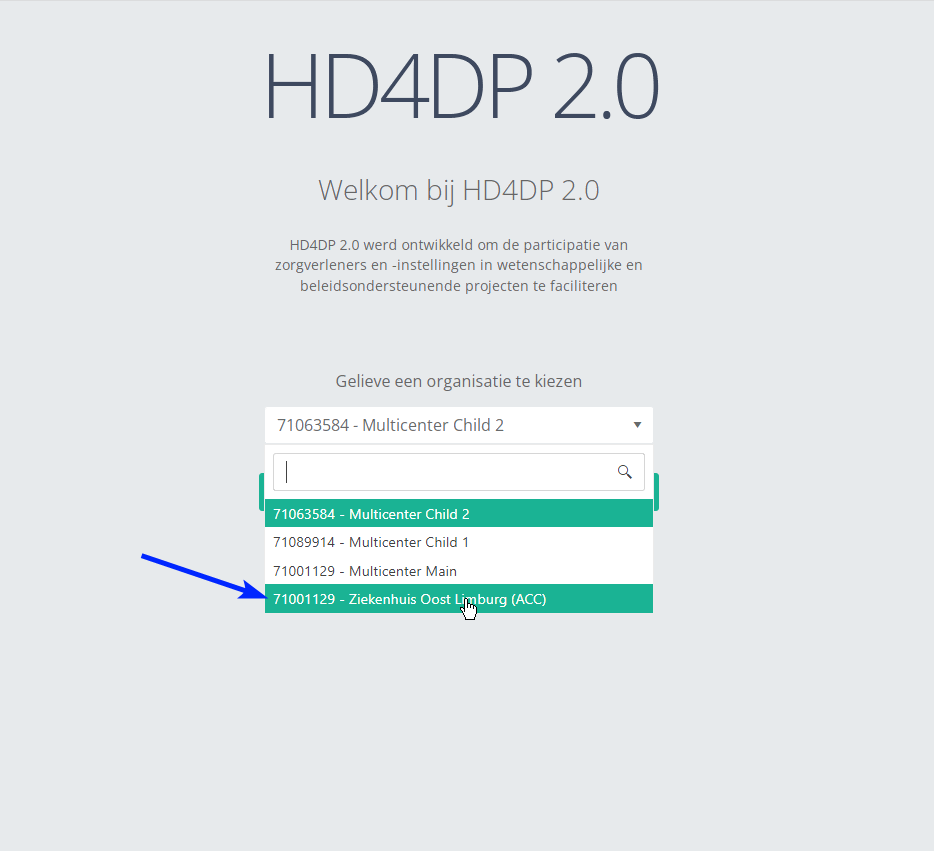
Step 2: Select the concerned organization in the dropdown list and click on Volgende (Next)
Step 3: Fill in the username and password, that has been provided by your IT Department or Healthdata team, and click on Log in to access the HD4DP 2.0 application.
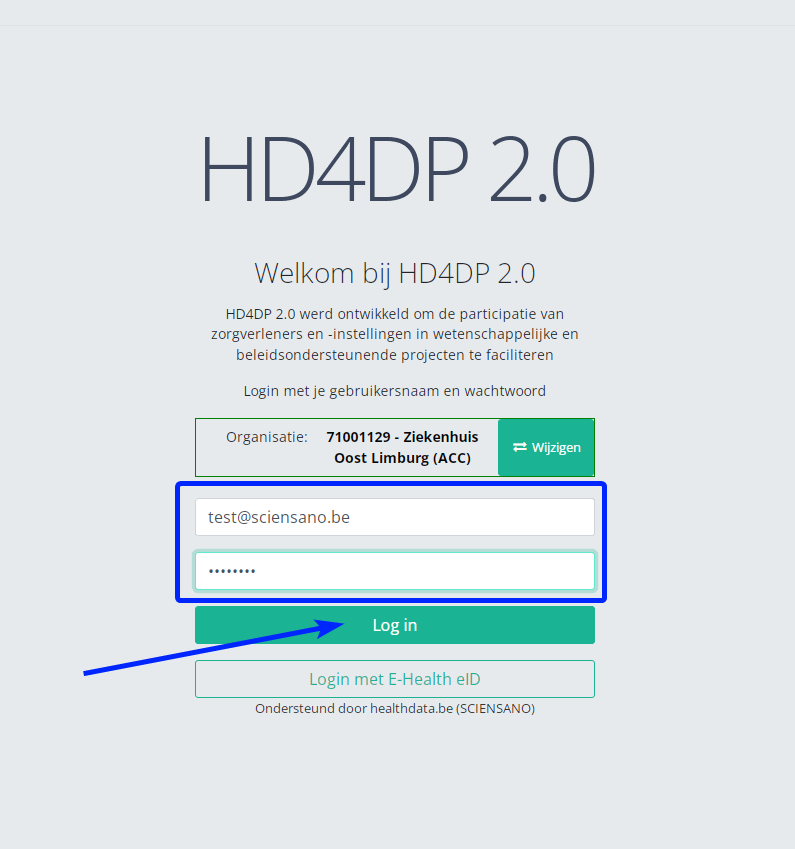
Step 4: Navigate in the menu on the left-hand side panel to the desired study program:
<Screenshot of the menu item on the left-hand side panel>
Step 5: Check that the uploaded registration(s) is/are displayed in the overview table:
<Screenshot of registration on right-hand side panel>
HD4DP v2 CSV Upload
HD4DP v2 CSV UploadThe CSV upload functionality enables the import of multiple parameters from a set of patients in one go into HD4DP2.0. The CSV file is based on an extract of the electronic patient files and/or other local databases.
Currently there is no user interface in HD4DP2.0 for the upload of CSV files. If a data provider wants to upload a CSV file, it has to be dropped at a specific location. These files will be picked up and processed periodically. The files should be “final”, meaning that no application is writing to them. The pick-up location will be identical for all registries.
Pre-registry handling will be based on a naming convention of the CSV file.
Refer to the page(s) hereunder for DCD specific CSV Upload documentation.
CSV Upload for IQEDFOOT
CSV Upload for IQEDFOOTDocumentation for CSV Upload on Architecture 2.0
Description of the service
The CSV Upload functionality gives the possibility to import multiple parameters from a set of patients in one time into HD4DP 2.0. The csv file is based on an extract of the electronic patient files and/or other local databases.
Currently there is no user interface in HD4DP 2.0 for uploading csv files. If a data provider wants to upload a csv file, the file has to be dropped at a specific location. These files will be picked up and processed periodically. The files should be “final”, meaning that no application is writing to them. The pick-up location will be identical for all registries.
Pre-registry handling will be based on a naming-convention of the csv file.
HOW TO: Upload data using CSV Upload
Steps To Upload data
1. Prepare the csv file (example file in this section)
- Extract the csv file from the electronic patient files and/or other local databases.
- Author group, Author and Coauthor:
- When the Author Group, Author and Coauthor have been left out in the csv file, the default Author group, Author and Coauthor will be used automatically.
- When the desired Author Group, Author and Coauthor are specified in the csv file, the following headers TX_AUTHOR_GR, TX_AUTHOR and TX_COAUTHOR must be added to the csv file with their values respectively.
Example:
TX_AUTHOR_GR;TX_AUTHOR;TX_COAUTHOR
Test group;test@sciensano.be;test@sciensano.beNote:
The Author group, Author and Coauthor must exist and are well configured at the back-end of the system. TX_AUTHOR_GR can be a string that identifies the Author group to which this Author belongs. Commonly, the first name and last name are used to identify the TX_AUTHOR_GROUP. Be sure to avoid leading and trailing spaces when entering the Author group value.
- Similarly, add the field name 'STATUS' in capitals in an additional column. Add the value 'draft' in case a manual submission of the record is requested.
If not, the record will be submitted without manual intervention.
- Adding separators to a NISS number:
It is not necessary to add separators in a NISS number when uploading a file using CSV Upload. You can fill out the NISS number both with or without separators. E.g.: 85.04.02-169.32 or 85040216932.
Example:
- Make sure the name of the csv file has the correct format:
HD_DCD_submcsv_HDBPnumber_HDBPabbreviation_versionnumber_versionreleasedate
- Make sure the name of the csv file has the correct format: HD_DCD_submcsv_HDBPnumber_HDBPabbreviation_versionnumber_versionreleasedate
So for <dcd name> the format would be : HD_DCD_submcsv_<HDBPnumber>_<HDBPabbreviation>_<versionnumber>_<versionreleasedate>
Please find here an example file for <dcd name>:
<paste the content of the csv file here>Disclaimer: The example files above are only provided as a guideline and do not contain real life data.
2. Uploading the csv file
Step 1: Open the sftp tool like WinSCP

Step 2: Get the credentials (Host name, Port number, User name and Password) from the IT department of the Data Provider, to log on to the sftp server located on the Data Provider side. See section Data transfer via an SFTP client in the documentation of the Online Acceptance Environment.
Step 3: Fill in the credentials into the Login screen and click on Login to be able to access the different upload folders:
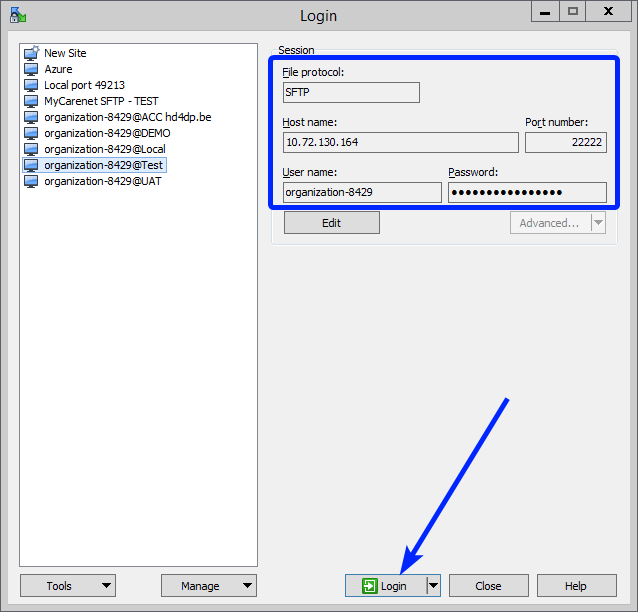
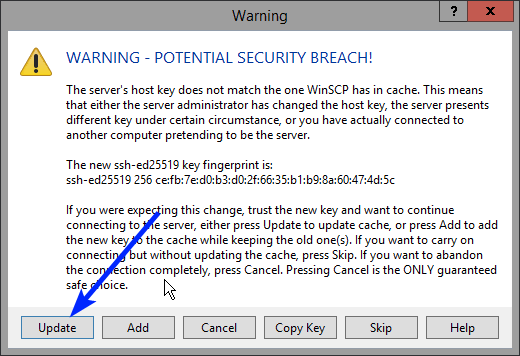
Note: a warning might be given, just click on Update
Now the CSV Upload folder structure is displayed on the right-hand side panel. You will notice draftcsv folders and submcsv folders for the diverse HD business projects. Historically, a directly submitted csv file would end up in the submitted folder, whereas a csv file featuring ‘draft’ in the status field would have ended up in the draft folder. At this stage there is no real distinction between both folders, so you can save the csv file in either of them.
Attention: If you wish to create a draft to work on at a later stage, don’t forget to add ‘draft’ in the status column of the csv file.
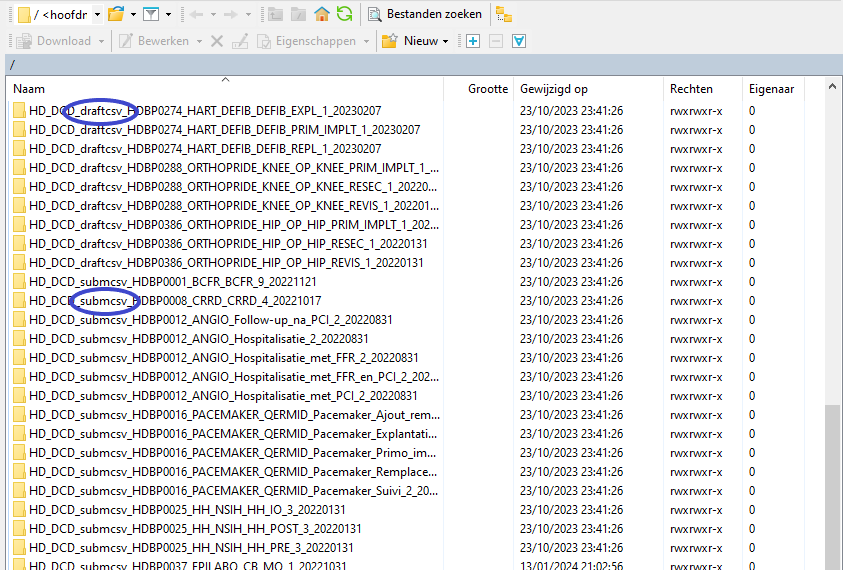
Step 4: Select the project folder <project folder name> and open it by double-clicking on it:
<Screenshot of Project with DCD folders>
Step 5: Double-click on the DCD folder to open it:
<Screenshot of DCD folders>
Step 6: Now go to the folder on the left-hand side panel where the csv file to be uploaded is located:
<Screenshot of DCD folders>
Step 7: Drag the csv file to be uploaded from the left-hand side panel into the folder on the right-hand side panel:
<Screenshot of file on right-hand side panel>
Step 8: Wait until the polling system of the CSV Uploader has picked up the CSV file and has processed it.
Once the csv file has been processed it will disappear from the folder, when we refresh the page manually!
<Screenshot of DCD folder where the file has disappeared after a refresh>
3. Validate csv upload
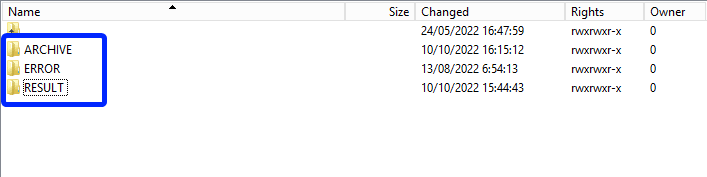
Once the csv file has been processed 3 folders will be created (if they haven't been created already) in the DCD folder:
- ARCHIVE (after a csv file has been processed, the original csv file will be saved in this folder)
- RESULT (when the csv file is placed in this folder, it means that the csv file has been processed, a file will be created or (or appended, if the file already existed) with the result of the upload of the csv file).
All the errors that are described in this file are business related, which means that they are technically correct, but in violation with the business rules or contain wrong values for that field. - ERROR (when the csv file is placed in this folder, it means that a technical error has occurred like the csv file contained erroneous formatting. The csv file won't get processed and an error file will be created with the errors and reason why the csv file couldn't be processed)
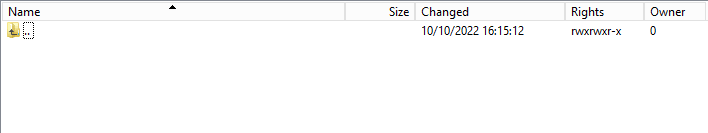
3.1 Validation of the csv upload via sftp tool:
Step 1: Double-click on the ERROR folder to open it, click on the refresh button and verify that there is no error file present.
Step 2: Return to the DCD folder. Now double-click on the RESULT folder to open it, click on the refresh button and verify that the result file is present.
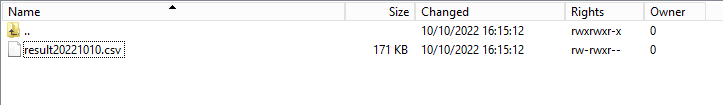
Step 3: Double-click on the result file to open it.
<Screenshot of result file of the DCD>
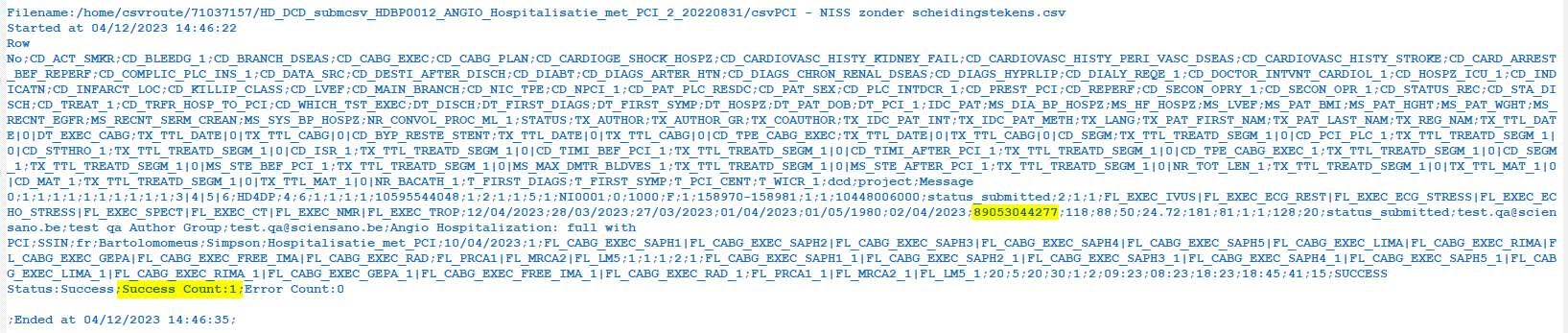
Step 4: If there are multiple records in the result file, scroll to the entry of the current csv upload by looking at the upload date (Started at dd/mm/yyyy hh:mm).
Verify the result file that the upload was successful by searching for the word SUCCESS and having a look at the Status. This Status must contain: Success;Success Count:1;Error Count:0
<Screenshot of result file of the DCD>
3.2 Validation of the csv upload via HD4DP 2.0:
Step 1: Open the web application HD4DP 2.0.

Step 2: Select the concerned organization in the dropdown list and click on Volgende (Next)

Step 3: Fill in the username and password, that has been provided by your IT Department or Healthdata team, and click on Log in to access the HD4DP 2.0 application.

Step 4: Navigate in the menu on the left-hand side panel to the desired study program:
<Screenshot of menu navigation to the DCD>
Step 5: Check that the uploaded registration(s) is/are displayed in the overview table.
<Screenshot of DCD Overview screen>
Databases
Databases Bart.Servaes Fri, 02/16/2024 - 10:04Retrieve IQEDFOOT data from the local database of HD4DP v2
Retrieve IQEDFOOT data from the local database of HD4DP v2Warning
The person with the login for the local database of "HD4DP v2 local" has access to all the data stored in the database. This means that the personal data of the patients will be VISIBLE to that user.Requirements
URL Local DWH Database: postgresql://<server_ip>:5432/localdwh. If this is not the case, the IT department hosting HD4DP v2 needs to open the port and allow traffic to this port.
URL NIPPIN Database: postgresql://<server_ip>:5432/nippin
Username/Password: The service desk of healthdata.be will forward, via a secure link, the username and password.
Client: Download one of the clients that support PostgreSQL . A list is available here.
IP network/subnet: Provide us with the IP network/subnet from where you will contact the database. The database only accepts incoming traffic of known IP networks/subnets. example: 0.0.0.0/32
Granted privileges
| database | user | privileges |
| localdwh | dpuser | CONNECT/local_dwhmessage:SELECT/local_dwhmessage_key_value:SELECT/local_dwhmessage_key_value_plus:SELECT |
| nippin | dpuser | CONNECT/nippin_message:SELECT/nippin_cleanup:SELECT |
"data_collection_name" in local database
- "data_collection_name" =
Query examples
With the "data_collection_name" and the following information, you will be able to link multiple tables with each other.
- local_dwhmessage_key_value: Key value table with more information about the registration
- msg_document_id: document id of your message located in local_dwhmessage table
- document_id: document id of your registration
- local_dwhmessage: table where you can find all the registrations
- local_dwhmessage_key_value_plus: Extra table to define attribute type and value of a key value
- key_value_id: Key value id linked to the id of the local_dwh_message_key_value
- local_dwhmessage_key_value:
"local_dwhmessage_key_value" column "msg_document_id" refer to the "document_id" of "local_dwhmessage".
"local_dwhmessage_key_value_plus" column "key_value_id" refer to the id of "local_dwhmessage_key_value".
Query 1: Get all registrations from the last 15 days.
SELECT * from local_dwhmessage WHERE data_collection_name = 'add data_collection_name' and created_on > current_date - interval '15' day;Query 2: Get all registrations and key value.
SELECT * from local_dwhmessage as ldm left join local_dwhmessage_key_value as ldmkv on ldmkv.msg_document_id = ldm.document_id WHERE ldm.data_collection_name = 'add data_collection_name';Query 3: Get all registrations, key value and key value plus from.
SELECT * from local_dwhmessage as ldm left join local_dwhmessage_key_value as ldmkv on ldmkv.msg_document_id = ldm.document_id left join local_dwhmessage_key_value_plus as ldmkvp on ldmkvp.key_value_id = ldmkv.id WHERE ldm.data_collection_name = 'add data_collection_name';This documentation is being updated regularly. We try to provide as correct, complete and clear as possible information on these pages. Nevertheless, if you see anything in the documentation that is not correct, does not match your experience or requires further clarification, please create a request (type : request for information) via our portal (https://sciensano.service-now.com/sp) or send us an e-mail via support.healthdata@sciensano.be to report this documentation issue. Please, do not forget to mention the URL or web address of the page with the documentation issue. We will then adjust the documentation as soon as possible. Thank you!Architecture 2.5
Architecture 2.5 Bart.Servaes Wed, 10/25/2023 - 16:02Uniformed naming convention
Uniformed naming conventionChange name:
Create DCD formio naming structure using a general overall naming convention of healthdata projects.
Change Description:
The change is to refactor the formio DCD naming to have a structure as:
HDBPnumberHDBPabbreviationdcdname/abbreviationversionnumber
Where the abbreviation will be used as seen in the column “TX_PROJ_BUS_ABBREV” of the document below, that will be a reference to fill the MDM:
Example 1:
So for the formio DCDs of the project “Mandatory registration of spine surgery” (full name) it would be something like:
HDBP0240_SPINE_1 and because there is only 1 dcd for the project we do not use a dcd name.
Example 2:
So for the formio DCDs of the project “Pneumology Endobronchial valve” (full name) it would be something like:
HDBP0231_ZEPHYR_ZEPHYR_REPLAC_1
HDBP0231_ZEPHYR_ZEPHYR_PRIM_IMPLT_1
Full example:
dcdname/abbreviation: will be the TX_REG_NAME of the DCD.
| form title | form name | form path | collection name |
| HDBP0240_SPINE | hdbp0240Spine1 | hdbp0240spine1 | HDBP0240SPINE1 |
| HDBP0231_ZEPHYR_ZEPHYR_PRIM_IMPLT | hdbp0231ZephyrZephyrPrimImplt1 | hdbp0231zephyrzephyrprimimplt1 | HDBP0231ZEPHYRZEPHYRPRIMIMPLT1 |
Sending code values instead of code IDs in S2S requests
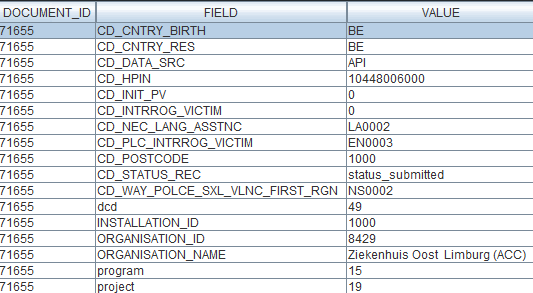
Sending code values instead of code IDs in S2S requestsIn the following example we will demonstrate sending code values instead of code IDs. We want, for instance, to send a patient's country and zip code information, more specifically for someone who lives in Brussels, Belgium:
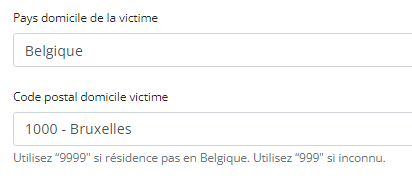
Currently, in the case of fields containing code list elements with S2S submissions, we send the code ID in the body of the message, like so:
"CD_CNTRY_RES": "130349",
"CD_POSTCODE": "4525",In CSV upload, however, we send the code value of the code object.
From the new Architecture 2.5 onward, we will do so too for S2S submissions using the API. So, for the example above, the new way of sending the same values will be as follows:
"CD_CNTRY_RES": "BE", "CD_POSTCODE": "1000",The image below shows the relationship between these fields in the database, in the case of the country:

And for the postal code:

The purpose of this change is to stop sending the code IDs in System-to-System and start sending the code values, just like we are doing in the CSV upload. The performance improvements that go along with this result from the fact that the service that returns the code value for a given ID is done through a single request per submission to the MDM, thus saving a lot of time in calls to get these values.
Since the code value is not unique, we need to get the codelist ID of an element, given its key, from a given DCD version ID. This could be summarized for this example as follows:
The key CD_CNTRY_RES on DCD version ID 55 (BSACC Police And Public Prosecutor) has the codelist id 347.
The key CD_POSTCODE on DCD version ID 55 (BSACC Police And Public Prosecutor) has the codelist id 12.Code Lists and Code Values are agreed upon by IAT and the Researchers. These are sent to DC by IAT via the DCD specifications. A Code ID is the identification of a Code Value in MDM. So how is the translation done?
In the case of sending code IDs, the user calls our services to retrieve the code IDs through requests to our API. In these requests, the user always receives the complete code object, containing not only the code ID, but also the code value. Let’s have a look at an example for the country code (code list 347):
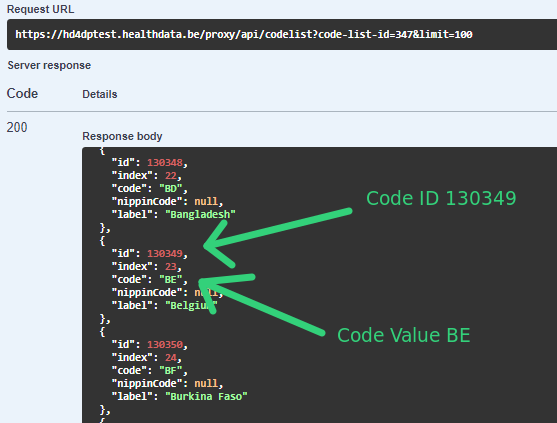
The image shows how we were sending the code ID 130349 in System-to-System and how we can send the code value “BE” in Architecture 2.5, the same way that was done already using the CSV Uploader.
In terms of code, if we were already calling the services to get the code ID, the only thing that is necessary is changing the returned value from CodeID to CodeValue from the same Code object that is returned.
FROM:
mappedCode = codeFieldService.getFieldCode(field.getFieldId(), codeContent).codeId();TO:
mappedCode = codeFieldService.getFieldCode(field.getFieldId(), codeContent).codeValue();That’s the only change needed in the mapping. All the rest remains exactly the same.
For more information, please check our documentation at 1. End-to-End process to submit DCD registrations | docs.healthdata.be
Architecture 2.0 and Architecture 2.5 proxies
Below you find an example of how the service to get a project-result is called in both proxies:
Architecture 2.0
https://hd4dp.acceptance.healthdata.be/proxy/api/installation/project-result?organization-id=120
[{"projectId":4,"dcdId":9,"dcdVersionId":9,"version":1,"formioName":"hdbp0231ZephyrZephyrPrimImplt1"}, {"projectId":4,"dcdId":10,"dcdVersionId":10,"version":1,"formioName":"hdbp0231ZephyrZephyrReplac1"}, {"projectId":4,"dcdId":11,"dcdVersionId":11,"version":1,"formioName":"hdbp0231ZephyrZephyrFllwup1"}, {"projectId":5,"dcdId":12,"dcdVersionId":12,"version":3,"formioName":"hdbp0062BelPstSpineTangoIntake1"}, {"projectId":5,"dcdId":13,"dcdVersionId":13,"version":3,"formioName":"hdbp0062BelPstSpineTangoConservativeTherapy1"}, {"projectId":5,"dcdId":14,"dcdVersionId":14,"version":3,"formioName":"hdbp0062BelPstSpineTangoPatientQuestionnaire1"}, {"projectId":5,"dcdId":15,"dcdVersionId":15,"version":3,"formioName":"hdbp0062BelPstSpineTangoSurgery1"}, {"projectId":6,"dcdId":16,"dcdVersionId":16,"version":1,"formioName":"hdbp0000TestTestDcd011"}, {"projectId":6,"dcdId":17,"dcdVersionId":17,"version":1,"formioName":"hdbp0000TestTestDcd021"}, {"projectId":7,"dcdId":18,"dcdVersionId":18,"version":1,"formioName":"hdbp0386OrthoprideHipOpHipPrimImplt1"}, {"projectId":7,"dcdId":19,"dcdVersionId":19,"version":1,"formioName":"hdbp0386OrthoprideHipOpHipRevis1"}, {"projectId":7,"dcdId":20,"dcdVersionId":20,"version":1,"formioName":"hdbp0386OrthoprideHipOpHipResec1"}, {"projectId":8,"dcdId":21,"dcdVersionId":21,"version":1,"formioName":"hdbp0288OrthoprideKneeOpKneePrimImplt1"}, {"projectId":8,"dcdId":22,"dcdVersionId":22,"version":1,"formioName":"hdbp0288OrthoprideKneeOpKneeRevis1"}, {"projectId":8,"dcdId":23,"dcdVersionId":23,"version":1,"formioName":"hdbp0288OrthoprideKneeOpKneeResec1"}, {"projectId":9,"dcdId":24,"dcdVersionId":24,"version":1,"formioName":"hdbp0048OrthoprideMegaOpMpPrimImplt1"}, {"projectId":9,"dcdId":25,"dcdVersionId":25,"version":1,"formioName":"hdbp0048OrthoprideMegaOpMpRevis1"}, {"projectId":9,"dcdId":26,"dcdVersionId":26,"version":1,"formioName":"hdbp0048OrthoprideMegaOpMpResec1"}, {"projectId":1,"dcdId":1,"dcdVersionId":30,"version":3,"formioName":"hdbp0025HhNsihHhPre3"}, {"projectId":1,"dcdId":2,"dcdVersionId":31,"version":3,"formioName":"hdbp0025HhNsihHhIo3"}, {"projectId":1,"dcdId":3,"dcdVersionId":32,"version":3,"formioName":"hdbp0025HhNsihHhPost3"}, {"projectId":10,"dcdId":27,"dcdVersionId":33,"version":1,"formioName":"hdbp0245TaviTaviImplt1"}, {"projectId":10,"dcdId":28,"dcdVersionId":34,"version":1,"formioName":"hdbp0245TaviTaviFllwup1"}, {"projectId":11,"dcdId":29,"dcdVersionId":35,"version":1,"formioName":"hdbp0000CorrectionForm1"}, {"projectId":13,"dcdId":34,"dcdVersionId":40,"version":2,"formioName":"hdbp0016PacemakerQermidPacemakerPrimoImplantation2"}, {"projectId":13,"dcdId":35,"dcdVersionId":41,"version":2,"formioName":"hdbp0016PacemakerQermidPacemakerAjoutRemplacementElectrode2"}, {"projectId":13,"dcdId":36,"dcdVersionId":42,"version":2,"formioName":"hdbp0016PacemakerQermidPacemakerRemplacement2"}, {"projectId":13,"dcdId":37,"dcdVersionId":43,"version":2,"formioName":"hdbp0016PacemakerQermidPacemakerExplantation2"}, {"projectId":13,"dcdId":38,"dcdVersionId":44,"version":2,"formioName":"hdbp0016PacemakerQermidPacemakerSuivi2"}, {"projectId":14,"dcdId":39,"dcdVersionId":45,"version":3,"formioName":"hdbp0019Bewsd3"}, {"projectId":15,"dcdId":40,"dcdVersionId":46,"version":2,"formioName":"hdbp0012AngioHospitalisatie2"}, {"projectId":15,"dcdId":41,"dcdVersionId":47,"version":2,"formioName":"hdbp0012AngioHospitalisatieMetPci2"}, {"projectId":15,"dcdId":42,"dcdVersionId":48,"version":2,"formioName":"hdbp0012AngioHospitalisatieMetFfr2"}, {"projectId":15,"dcdId":43,"dcdVersionId":49,"version":2,"formioName":"hdbp0012AngioHospitalisatieMetFfrEnPci2"}, {"projectId":15,"dcdId":44,"dcdVersionId":50,"version":2,"formioName":"hdbp0012AngioFollowUpNaPci2"}, {"projectId":16,"dcdId":45,"dcdVersionId":51,"version":1,"formioName":"hdbp0240Spine1"}, {"projectId":17,"dcdId":46,"dcdVersionId":52,"version":4,"formioName":"hdbp0008Crrd4"}, {"projectId":19,"dcdId":48,"dcdVersionId":54,"version":1,"formioName":"hdbp0242BsaccMeBsaccReprt1"}, {"projectId":19,"dcdId":49,"dcdVersionId":55,"version":1,"formioName":"hdbp0242BsaccMeBsaccFuPolce1"}, {"projectId":19,"dcdId":50,"dcdVersionId":56,"version":1,"formioName":"hdbp0242BsaccMeBsaccFuCont1"}, {"projectId":19,"dcdId":51,"dcdVersionId":57,"version":1,"formioName":"hdbp0242BsaccMeBsaccFuReferral1"}, {"projectId":19,"dcdId":52,"dcdVersionId":58,"version":1,"formioName":"hdbp0242BsaccMeBsaccFuMed1"}, {"projectId":19,"dcdId":53,"dcdVersionId":59,"version":1,"formioName":"hdbp0242BsaccMeBsaccFuPsychc1"}, {"projectId":20,"dcdId":54,"dcdVersionId":60,"version":9,"formioName":"hdbp0001Bcfr9"}, {"projectId":21,"dcdId":55,"dcdVersionId":61,"version":3,"formioName":"hdbp0078Pitter3"}, {"projectId":22,"dcdId":56,"dcdVersionId":62,"version":3,"formioName":"hdbp0051Becpr3"}, {"projectId":23,"dcdId":57,"dcdVersionId":63,"version":1,"formioName":"hdbp0037Epilabo1"}, {"projectId":24,"dcdId":58,"dcdVersionId":64,"version":1,"formioName":"hdbp0244RProfildRprofildTreat1"}, {"projectId":24,"dcdId":59,"dcdVersionId":65,"version":1,"formioName":"hdbp0244RProfildRprofildRenewal1"}, {"projectId":25,"dcdId":60,"dcdVersionId":66,"version":1,"formioName":"hdbp0274HartDefibDefibPrimImplt1"}, {"projectId":26,"dcdId":61,"dcdVersionId":67,"version":1,"formioName":"hdbp0056MvoMvoImp1"}, {"projectId":26,"dcdId":62,"dcdVersionId":68,"version":1,"formioName":"hdbp0056MvoMvoFu1"}, {"projectId":25,"dcdId":63,"dcdVersionId":69,"version":1,"formioName":"hdbp0274HartDefibDefibExpl1"}, {"projectId":25,"dcdId":64,"dcdVersionId":70,"version":1,"formioName":"hdbp0274HartDefibDefibRepl1"}, {"projectId":25,"dcdId":65,"dcdVersionId":71,"version":1,"formioName":"hdbp0274HartDefibDefibElect1"}, {"projectId":27,"dcdId":67,"dcdVersionId":72,"version":1,"formioName":"hdbp0000DvrForm1"}]Architecture 2.5
[{"projectId":19,"dcdId":49,"dcdVersionId":55,"version":1,"formioName":"hdbp0242BsaccMeBsaccFuPolce1"}]As we can see, using proxy (Architecture 2.0) we have all projects from organization 120, while proxy (Architecture 2.5) returns only one element in its response. This is because the DCD filter in proxy (Architecture 2.5) is done in the database more efficiently, while in the proxy Architecture 2.0 the same filter was done in the code after the service call.
So, the filter is not used in the first call and if Architecture 2.0 calls the same service in Architecture 2.5 without the filter (https://hd4dp.acceptance.healthdata.be/proxy/api/installation/project-result?organization-id=120) it will get exactly the same result.
Architecture 2.0 and Architecture 2.5 proxies tests
CSV Uploader
For the CSV Upload, nothing has changed. So the same inputs are valid for both architectures. Here are the example of some successful tests:
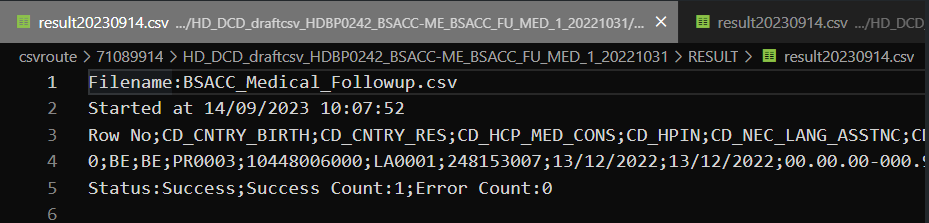
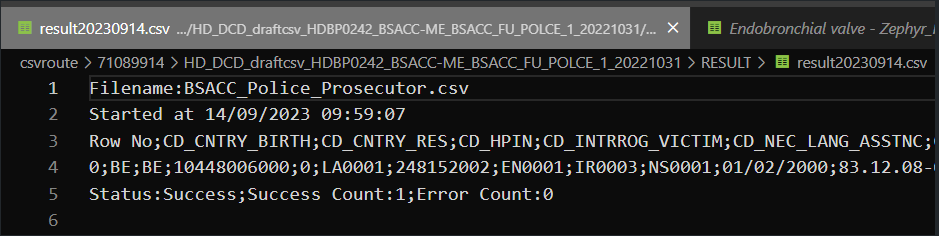
System-To-System
For S2S, as already mentioned, the only difference is that Architecture 2.0 sends Code IDs while Architecture 2.5 sends Code Values. Here are the same tests in both architectures:
Proxy (Architecture 2.0):
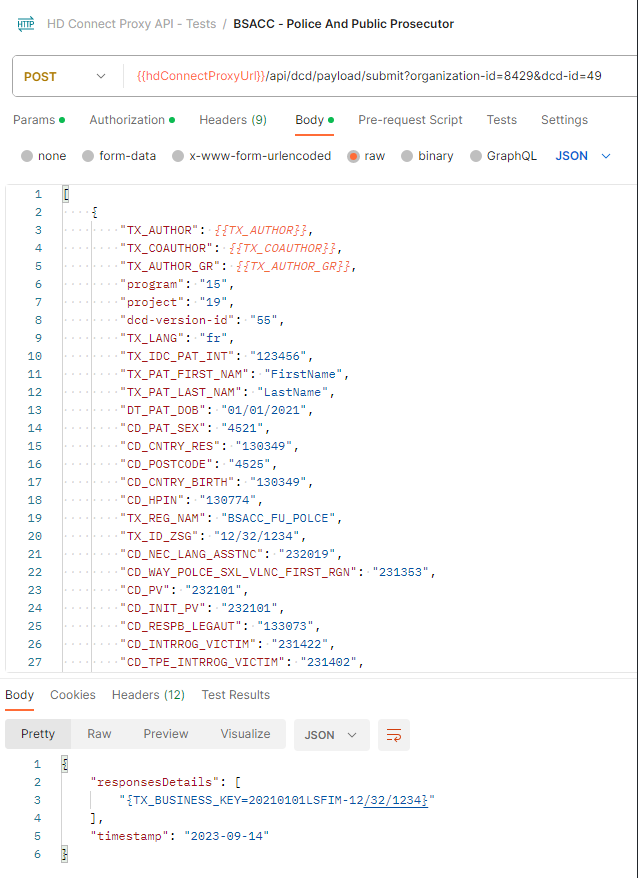
Proxy (Architecture 2.5):
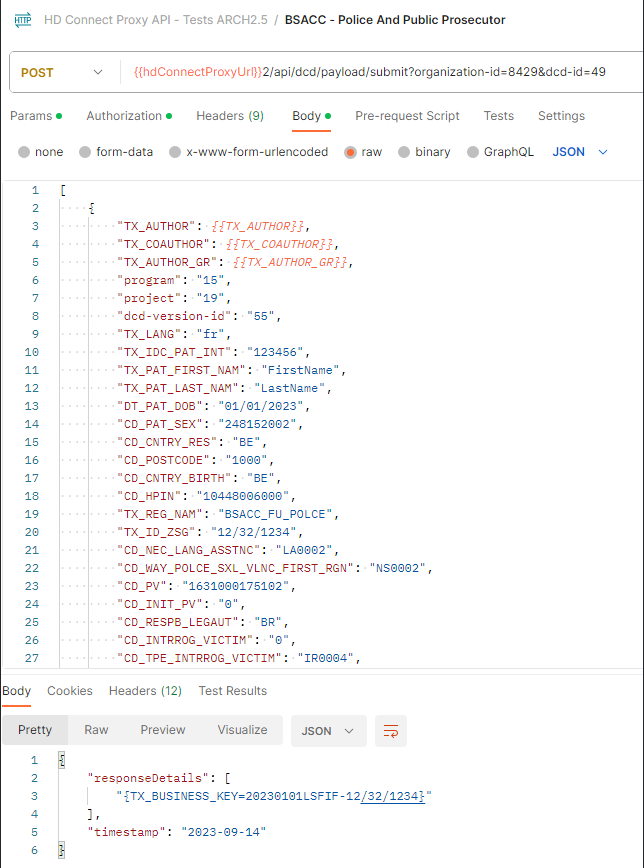
As we can see, we have the same request, where the first sends Code IDs and the second sends Code Values; both return a successful response.
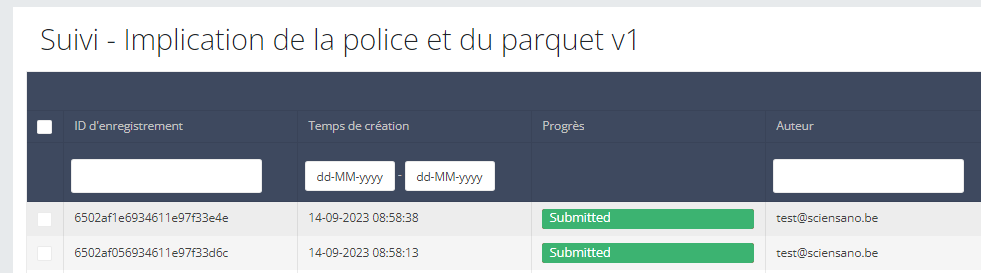
Front-end
In the front-end (GUI) everything is received normally as the same request was sent twice. After all, S2S will always send the same request with Code IDs to FormIO.
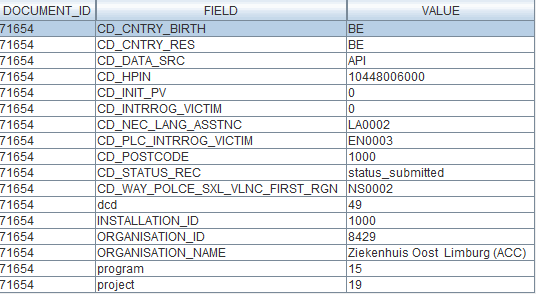
Data Warehouse
Once again, all the information arrives seamlessly, due to the fact that the FormIO validation already happened in the previous step:

First call (Architecture 2.0):
Second call (Architecture 2.5):
MDM Mapping of billing codes for MyCareNet
MDM Mapping of billing codes for MyCareNetNote: The below described mapping solution only applies to the study project of PACEMAKER.
In Architecture 2.0 the DCD fields related to billing codes for MyCareNet do not contain the billing code as their code value, instead this is just a numerical value, and the code value is only displayed in the label, e.g.:

When then sending key values to localdwh and dwh, the submission will contain a key value as follows:

When generating a message for MyCareNet, however, this same numerical value is used in the message payload. Keeping these numerical values, will generate MyCareNet messages with a incorrect billingcode, in this case <BillingCode>2</BillingCode>.

In Architecture 2.5 an MDM Mapping of billing codes for MyCarenet has been implemented. The value for the key will no longer be a numerical value such as 1,2,3… but an actual billing code. To facilitate this requirement, a new column has been introduced in the MDM to associate the billing code with the correct 13 character value, such as "182932-182943". This billing code will be accepted by MyCareNet.