HD4DP v2 (Health Data For Data Providers)
HD4DP v2 (Health Data For Data Providers)Welcome to the technical documentation pages for the component "Health Data for Data Providers v2, or HD4DP v2", provided by the service healthdata.be (Sciensano).
These pages provide information about the technical processes of the component. The following sections are (will be) provided:
- How to request access to an HD application for a certain project (EAM)
- Access the application HD4DP v2
- The description of the data tables in HD4RES v2 (documentation in progress)
This documentation is being updated regularly. We try to provide as correct, complete and clear as possible information on these pages. Nevertheless, if you see anything in the documentation that is not correct, does not match your experience or requires further clarification, please create a request (type : request for information) via our portal (https://sciensano.service-now.com/sp) or send us an e-mail via support.healthdata@sciensano.be to report this documentation issue. Please, do not forget to mention the URL or web address of the page with the documentation issue. We will then adjust the documentation as soon as possible. Thank you!General description of the application HD4DP v2
General description of the application HD4DP v2The HD4DP version 2.x Local is an electronic data capture (EDC) system: a computerized system designed for the collection of clinical data in electronic format for use in research supporting human public health policy. HD4DP (Health Data for Data providers) replaces the traditional paper-based data collection methodology and the proliferation of websites to streamline data collection and expedite the time to analysis and reporting.
Components and features
The HD4DP version 2.x Local application contains the following major components: NextGen Connect, Form.io, HD Connect (LOCAL Proxy), Local datawarehouse.
NextGen Connect
NextGen Connect is a health care integration engine that translates message standards into the standard required by the receiving system, including data formats and standards like HL7, DICOM, ANSI X12, ASCII, and XML. Main functionalities are filtering, transformation, extraction and routing.
The NextGen Connect component is used to handle all integrations within HD4DP 2.0 itself but also all integrations with the external world.
Data collections API: The form.io server offers a REST API which can be used to submit data for each known data collection. Data provider Master Systems cannot access this API directly but need to use the API exposed by the NextGen Connect component. This API is simply a proxy for the form.io API, but allows extra features on top of the form.io API such as security, monitoring, throttling, …
CSV API: For each data collection data can be submitted file-based using a CSV. A CSV can contain multiple data entries for a single data collection definition. These data entries are transformed and pushed by the NextGen Connect component towards the form.io server for potential manual post-processing and validation.
HL7 FHIR API: For some data collections an HL7 FHIR API will be available. The NextGen Connect component performs the transformation towards the Data Collections API and push the data into the form.io server.
Data delivery: the NextGen Connect component handles all routing of data towards the external world. This means it verifies the form.io server for completed data entries which have not yet been delivered. For each data entry that needs to be delivered, it determines where to send the data to, how it needs to be transformed and how it needs to be split. It performs all these actions in a guaranteed delivery fashion: it makes sure the data reaches its destination, possibly retrying when something went wrong.
Feedback: the NextGen Connect component coordinates the receival of feedback, potentially transforming it and pushing it towards the respective data collection entry using the data collections API.
Form.io
Form.io is a data management platform that includes a form builder with a drag and drop interface, management of data with complete API platform, management of users, offline forms, dynamic forms, automatic creation of API, and application embedding. In HD4DP v2, an Angular frontend application is available on top of the form.io server. This application provides a user interface to data providers in which they can see the different data collections for which they are allowed to record and submit data manually. A form.io backend server is responsible for providing the form definitions and registrations of new/updated entries.
HD Connect (LOCAL Proxy)
The HD Connect component is used to retrieve metadata from Master Data Management Database (MDM DB) residing on healthdata.be side.
Local datawarehouse
Each and every change in data entries on the form.io server is pushed towards the local datawarehouse (Local DWH) for easy reporting and data extraction. This local DWH consists of a PostgreSQL database.
Installation and maintenance
The application HD4DP v2 Local is provided without cost and installed remotely on the infrastructure of the healthcare organization by healthdata.be. Healthcare organizations are provided the system requirements for installation of HD4DP v2 application. Healthcare organizations that cannot provide the system requirements can opt to request access and use of a HD4DP v2 Local application of another healthcare organization. Healthcare organizations that cannot provide the system requirements and cannot access and use a HD4DP v2 Local application of another healthcare organization, can request access and use of HD4DP v2 WEB hosted by healthdata.be.
The application HD4DP v2 Local is maintained without cost remotely on the infrastructure of the healthcare organization by healthdata.be. The infrastructure on which the application HD4DP v2 Local is installed, should be maintained by the healthcare organization.
Typical use
- A Local Study Lead (takes responsibility for the study or project within the participating health care organization. Often this is the head of the clinical department involved in the study or project) can:
- Log in using active directory
- Grant access to Local Study Associate and Local Study Support
- Select and access project
- Create, Find, Update, Delete, Submit (towards healthdata.be) and Correct a registration using form.io component
- Create, Find, Update, Delete, Submit and Correct a registration using form.io component
- View all registrations for project
- A Local Study Associate (a health care professional that participates in the study or project) can:
- Log in using active directory
- Grant access to Local Study Support
- Select and access project
- Create, Find, Update, Delete, Submit (towards healthdata.be, MyCareNet and other destinations) and Correct a registration using form.io component
- View own registrations for project
- A Local Study Support (A Local Study Associate can delegate registration to a Local Study Support. Often this is an administrative collaborator or staff of a department medical coding) can:
- Log in using active directory
- Select and access project
- Create, Find, Update, Delete, Submit (towards healthdata.be, MyCareNet and other destinations) and Correct a registration using form.io component
- Can view own registrations for project
Position of HD4DP v2 in HD Architecture 2.0
Position of HD4DP v2 in HD Architecture 2.0The application HD4DP v2 is one of the components of the broader data collection and management architecture managed by the healthdata.be service of Sciensano. Below you can find a high level view of that data collection and management architecture (version 2).
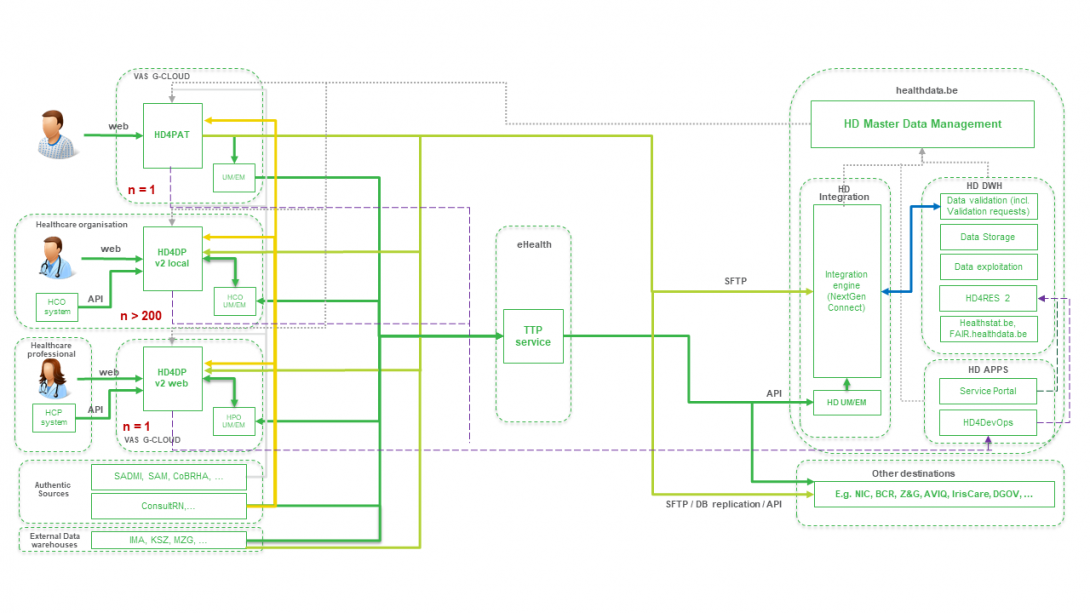
Dataflow description HD4DP v2
Dataflow description HD4DP v2Below we describe (high level) the dataflow between HD4DP v2 and the healthdata.be platform.
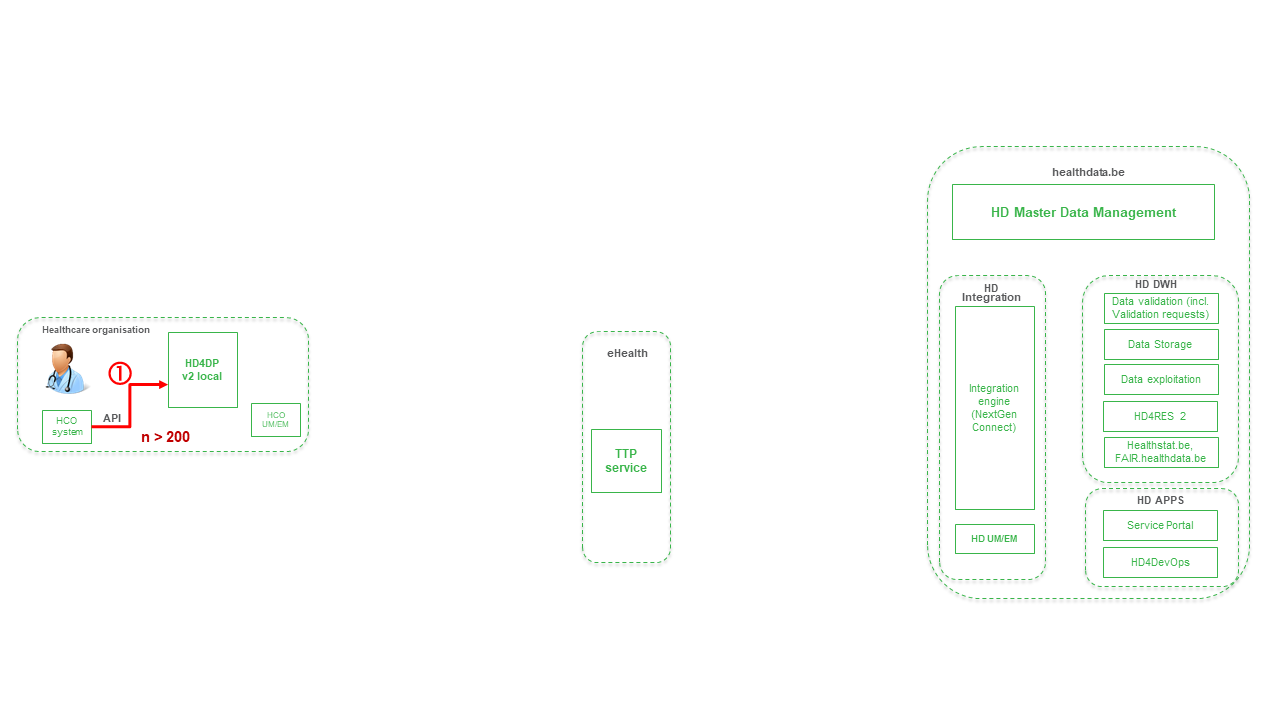
Step 1. Automatic data export from systems of data provider towards HD4DP v2 and prefill of forms if not complete.
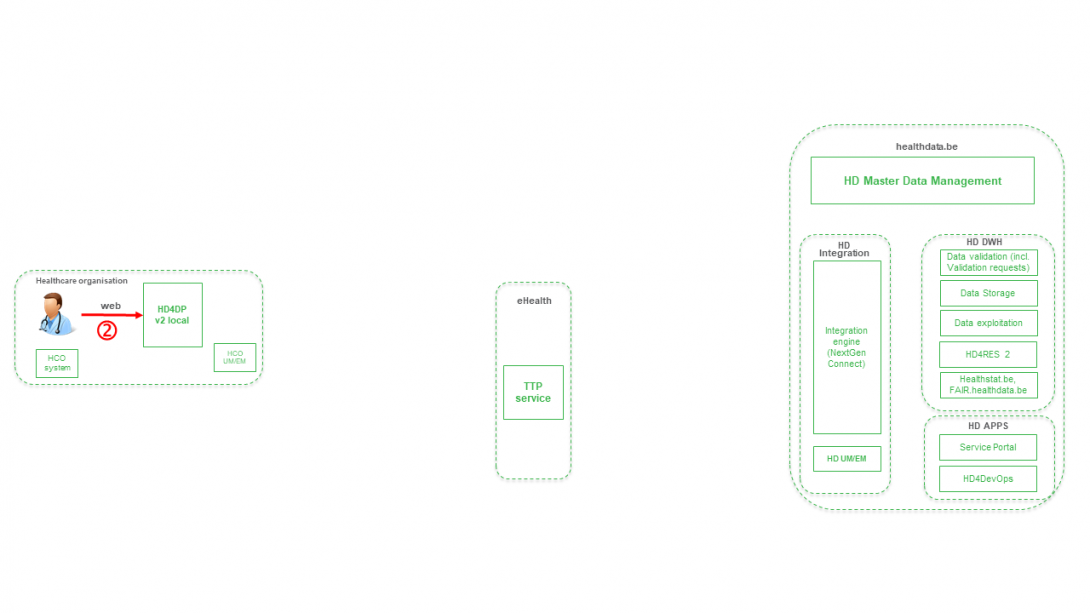
Step 2. Manual registration (de novo or completion) of data in form component of HD4DP v2.
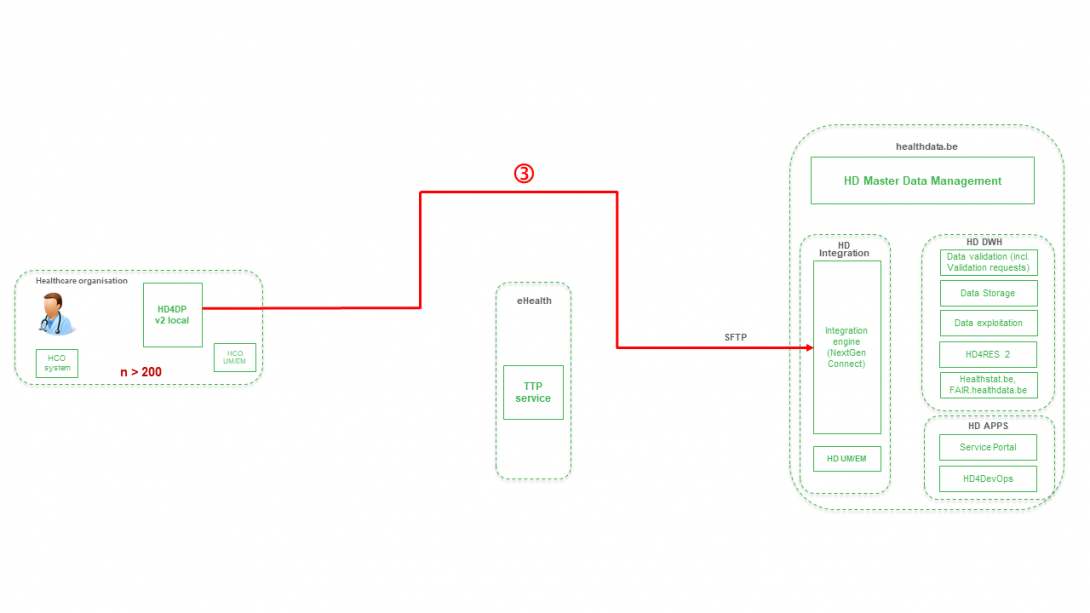
Step 3. Direct real time transfer of registry variables and technical ID of record from HD4DP v2 towards HD.
Step 4. Transfer of patient identifiers and technical ID of record from HD4DP v2 towards eHBox messaging client of HCO (HCO UM/EM).
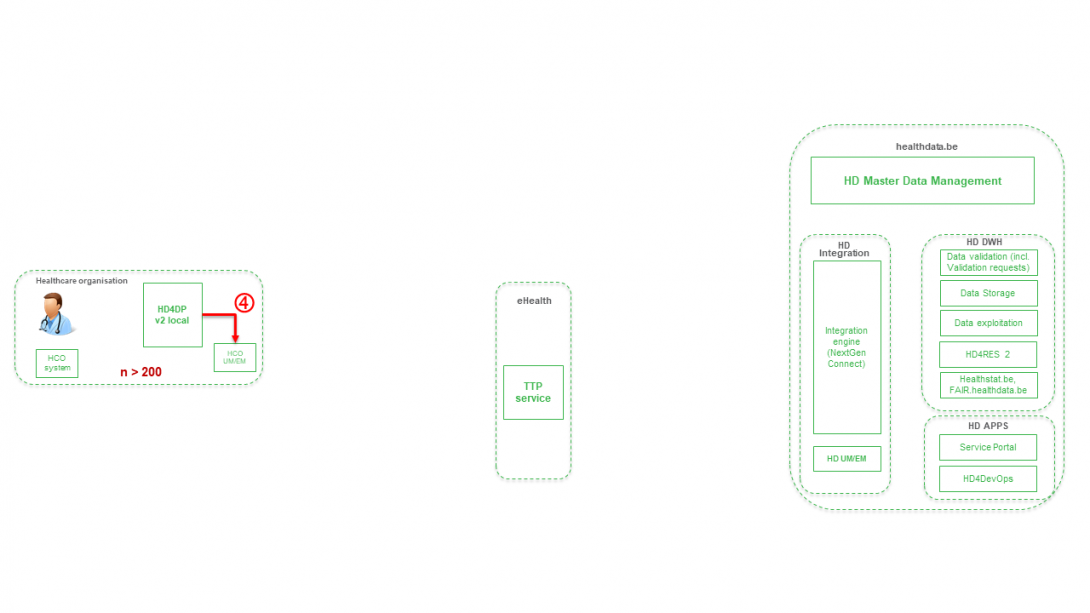
Step 5. Transfer of patient identifiers and technical ID of record from eHBox messaging client of HCO (HCO UM/EM) towards TTP service of eHealth.
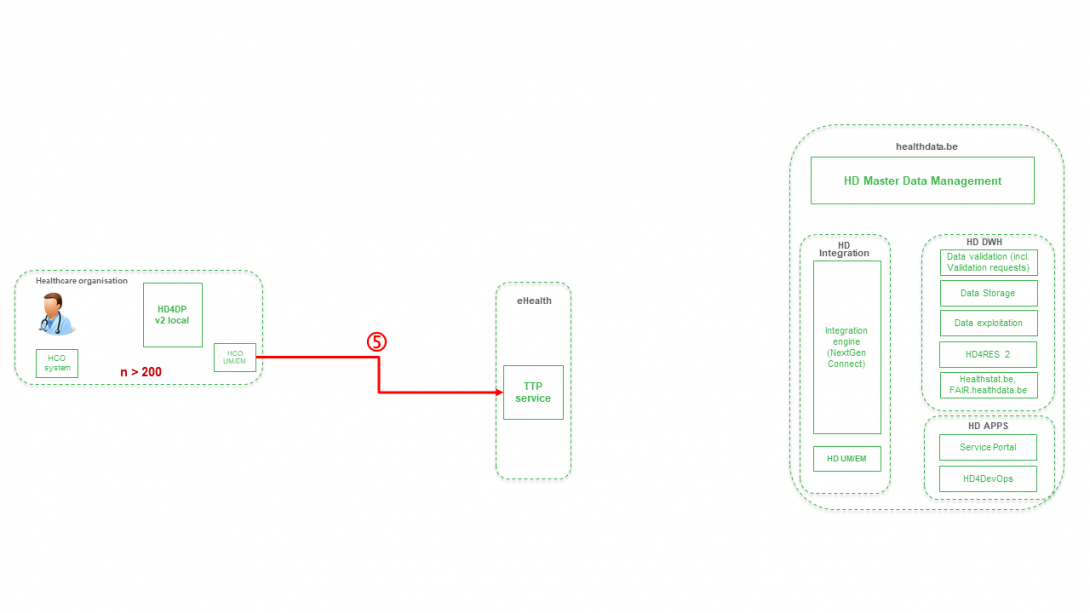
Step 6. Transfer of pseudonymized patient identifiers and technical ID of record from TTP service of eHealth towards eHBox messaging client of HD (HD UM/EM).
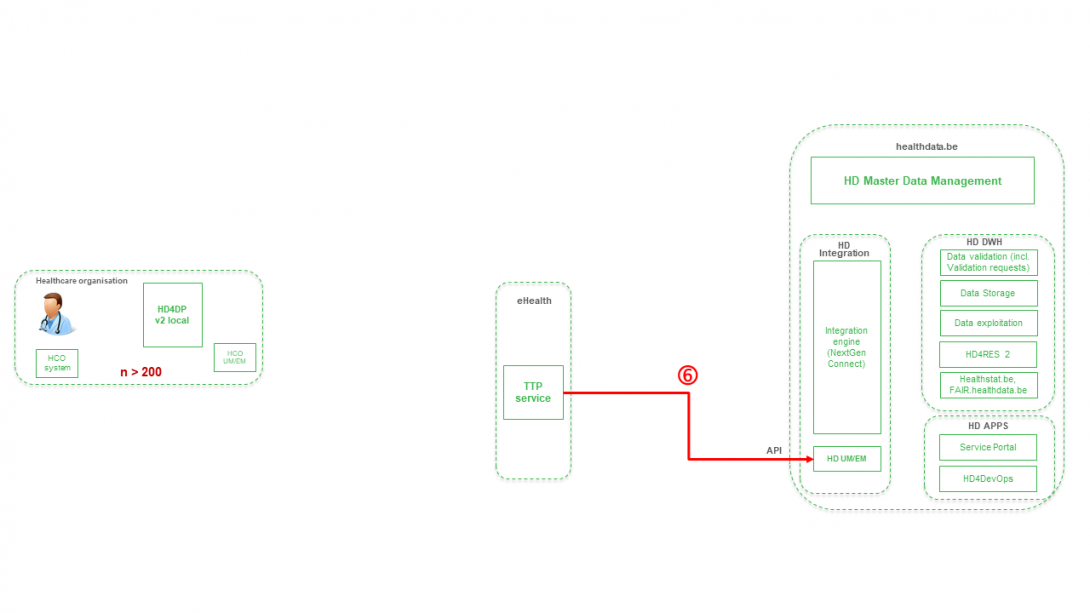
Step 7. Transfer of pseudonymized patient identifiers and technical ID of record from eHBox messaging client of HD (HD UM/EM) to HD Integration engine.
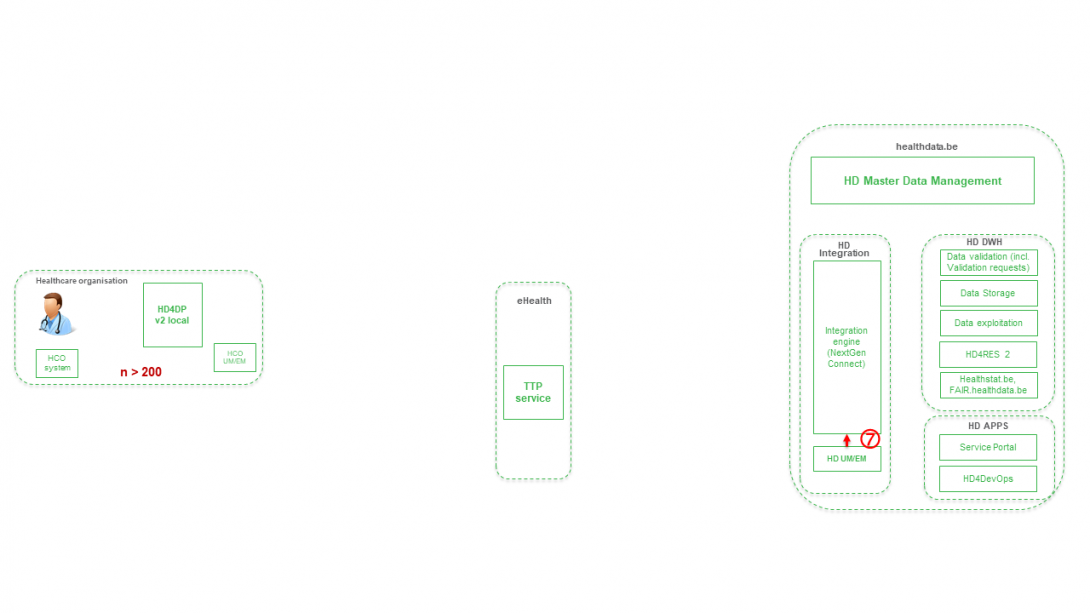
Step 8. Joining of pseudonymized patient IDs and the registry variables based on the technical ID and Transfer of the records from HD Integration engine towards Data Validation environment on DHW.
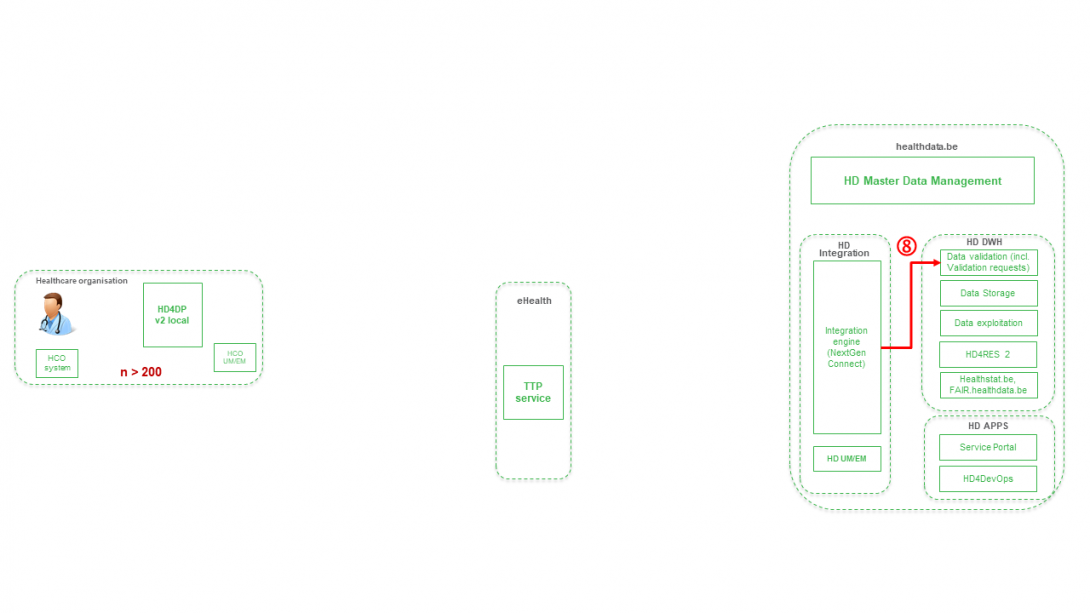
Step 9 - Option 1. Indirect transfer of patient identifiers and NIC-CIN variables from HD4DP v2 Local via the MyCareNet component of the HCO towards NIC-CIN (default).
Step 9 - Option 2. Direct real time transfer of patient identifiers and NIC-CIN variables from HD4DP v2 Local towards NIC-CIN (optional).
User manual of the application HD4DP v2
User manual of the application HD4DP v2In this manual we describe the following functions of the application HD4DP v2:
Request access to an HD application for a specific project
Request access to an HD application for a specific projectHealthdata.be applications such as HD4DP v2 and healthstat.be process sensitive personal information. Therefore, strictly controlled processes are used to grant access to these applications. The Entity Access Management (EAM) portal of healthdata.be facilitates these processes.
Due to the migration from EAM version 2.7 to EAM version 3.0 both systems will exist parallel during the transition period. Make sure that you always use the version currently available for your organization.
Below you will find the links to the user manuals to the different EAM systems. When selecting the desired version, the manual will appear in a separate tab.
When requesting access via the EAM portal version 2.7, go here.
When requesting access via the EAM portal version 3.0, go here.
This documentation is being updated regularly. We try to provide as correct, complete and clear as possible information on these pages. Nevertheless, if you see anything in the documentation that is not correct, does not match your experience or requires further clarification, please create a request (type : request for information) via our portal (https://sciensano.service-now.com/sp) or send us an e-mail via support.healthdata@sciensano.be to report this documentation issue. Please, do not forget to mention the URL or web address of the page with the documentation issue. We will then adjust the documentation as soon as possible. Thank you!User roles in HD4DP v2
User roles in HD4DP v2Each healthcare organisation has an Access Manager who watches over the access rights to the applications of the own organisation and manages them in the HD Entity Access Management (EAM) system. In this process access requests by healthcare organisation employees are analysed and validated if legitimised. The scope of the accesses granted may differ, which is reflected in various user roles. Based on access rights, following three user roles can be distinguished:
Local Study Lead (author):
The Local Study Lead can:
- edit and review all peer registrations (regardless of role) for the study or project;
- make registrations in HD4DP v2
This role might be but should not be limited to the responsible for the study or project within the participating healthcare institution.
Local Study Associate (author):
The Local Study Associate can:
- edit and review the own registrations, not those of other colleagues from the same healthcare organisation participating in the same study or project. The indicated registrations are limited to the patients treated by the Local Study Associate;
- make registrations in HD4DP v2.
The Local Study Associate is a healthcare provider participating in the study or project. This is reflected in the registration form.
Local Study Support (co-author):
The Local Study Support can:
- edit and review registrations belonging to the author group they are linked to;
- make registrations in HD4DP v2.
A Local Study Associate and Local Study Lead can delegate registration tasks to a Local Study Support. This might be but should not be limited to an administrative assistant or staff from a medical coding department. The Local Study Associate and Local Study Lead are still considered the author of the registration; the Local Study Support is considered the co-author. The Local Study Associate and Local Study Lead can view and modify Local Study Support entries.
By default, only 1 Local Study Lead is intended by healthdata.be (Sciensano) for each project within each organisation. The idea is that only one person is meant to see all submissions for that project within that organisation. This policy prevents users of HD4DP v2 from seeing personal and sensitive information from individuals with whom they do not have a therapeutic relationship. For policy deviations on this, healthcare organisation staff should contact their Data Protection Officer (DPO).
Remarks:
- The scope of the access rights does not necessarily reflect the hierarchy within your healthcare organisation.
- It is up to the Access manager to change roles from/to Local Study Lead, Local Study Associate and Local Study Support. These requests are carried out in the EAM system.
Access the application HD4DP v2
Access the application HD4DP v2To access the application HD4DP v2, you must first request an account. If you do not have an account yet, please consult "Request access to an HD application for a specific project" first.
Once your account has been created and the registry is put in production, you will receive an e-mail with the following information (Note that the text between the [ ] will be adapted):
- Organization: [RIZIV number - Name]
- Login: [email]
- Password: [password]
- Application URL: [url]
With these credentials you can access the application HD4DP v2 of your organization:
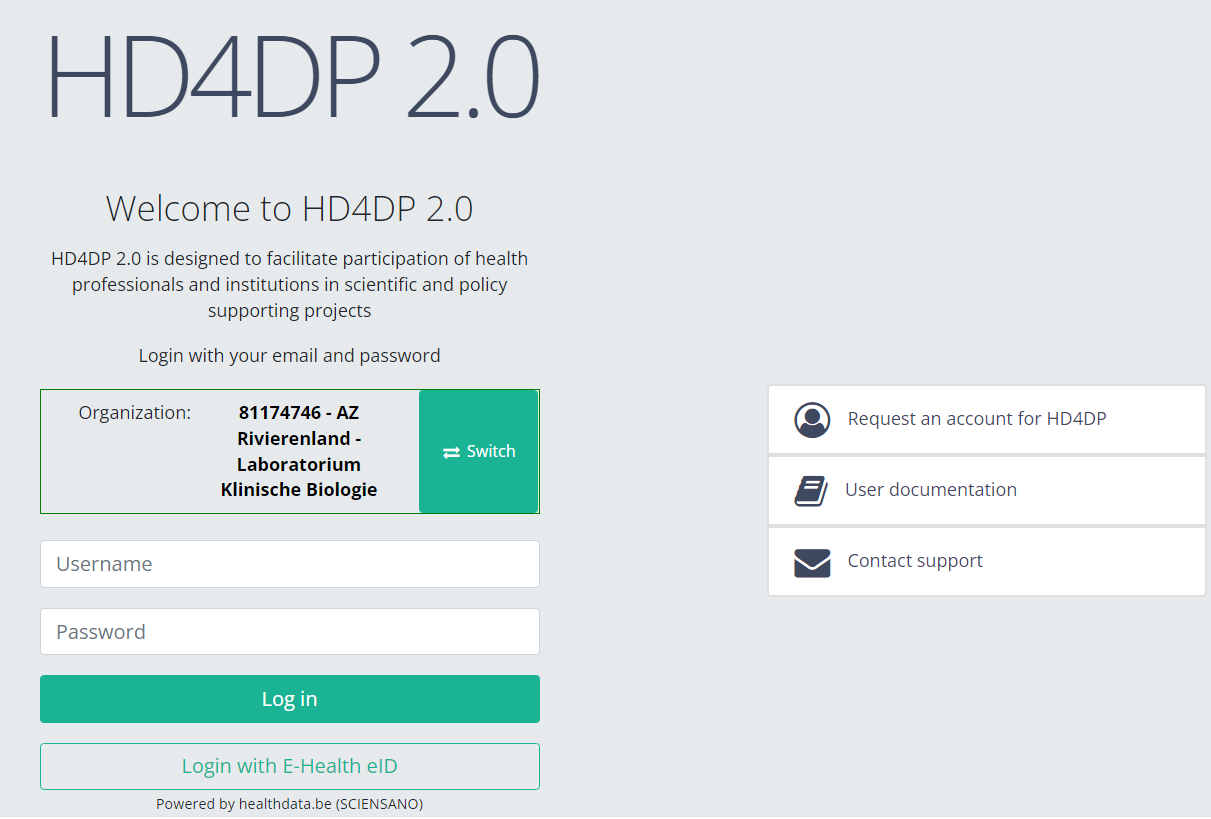
- Go to the url mentioned in the email
- Select "your organization" from the list
- Your organization: [RIZIV number – Name]
- Click on "Next"
- Fill in your "username" and "password"
- Click on "Log in"
Please be sure to log off after making use of the application HD4DP 2.0, or any other Healthdata.be application. Just closing your internet browser does not guarantee that your application with registrations has been closed.
Navigate to a project
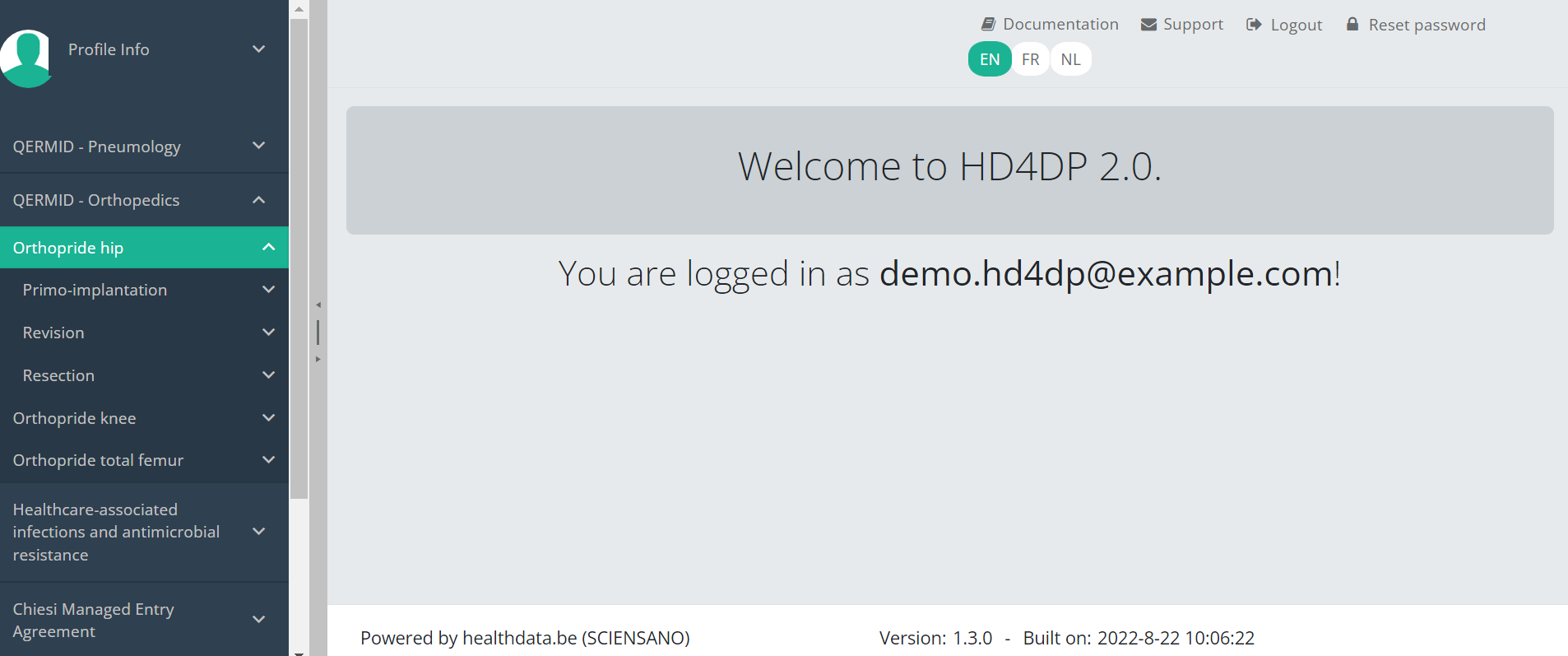
Navigate to a projectWhen logged in, you will see the Welcome page of HD4DP v2. In the left dark blue menu you can see all the study programs and projects you have access to.
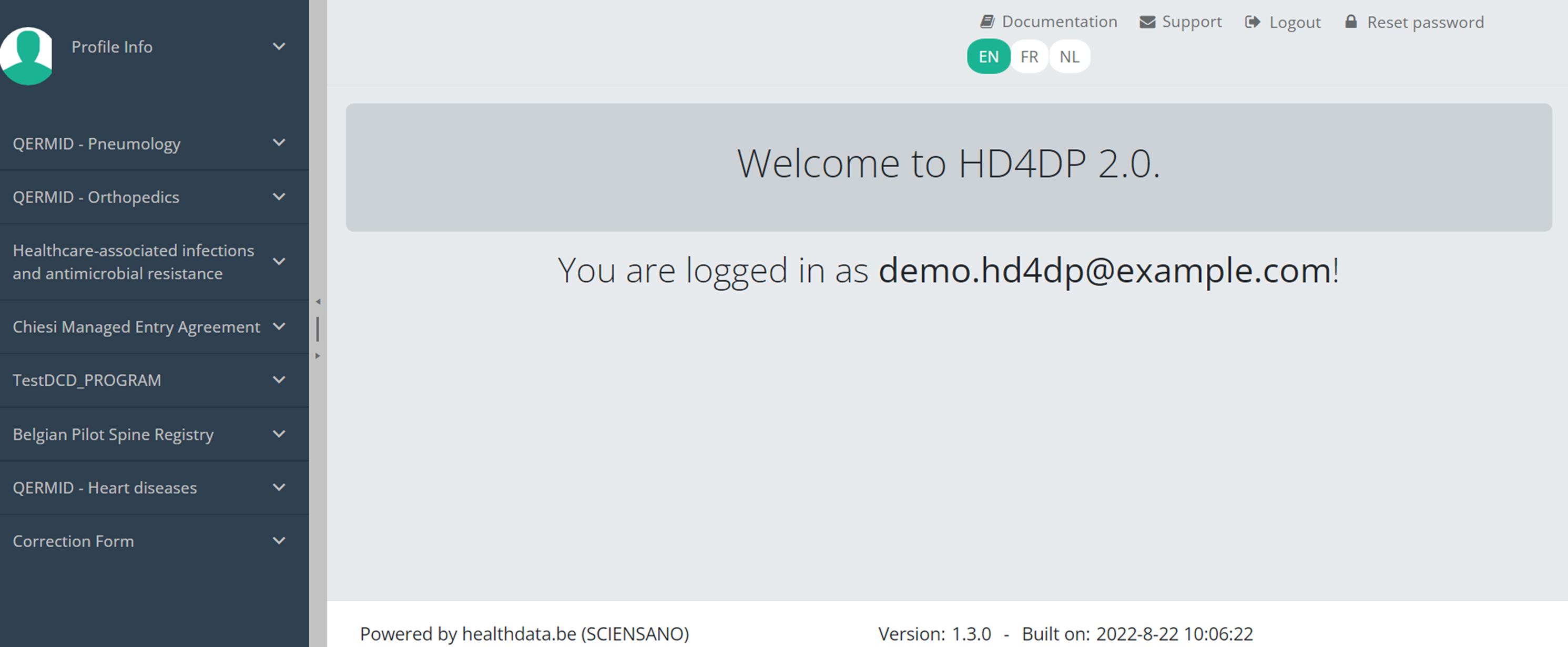
Suppose we want to participate in study Orthopride Hip. In that case, we must select the study program QERMID Orthopedics first. You can see now three study projects: Orthopride Hip, Orthopride Knee and Orthopride Total femur.
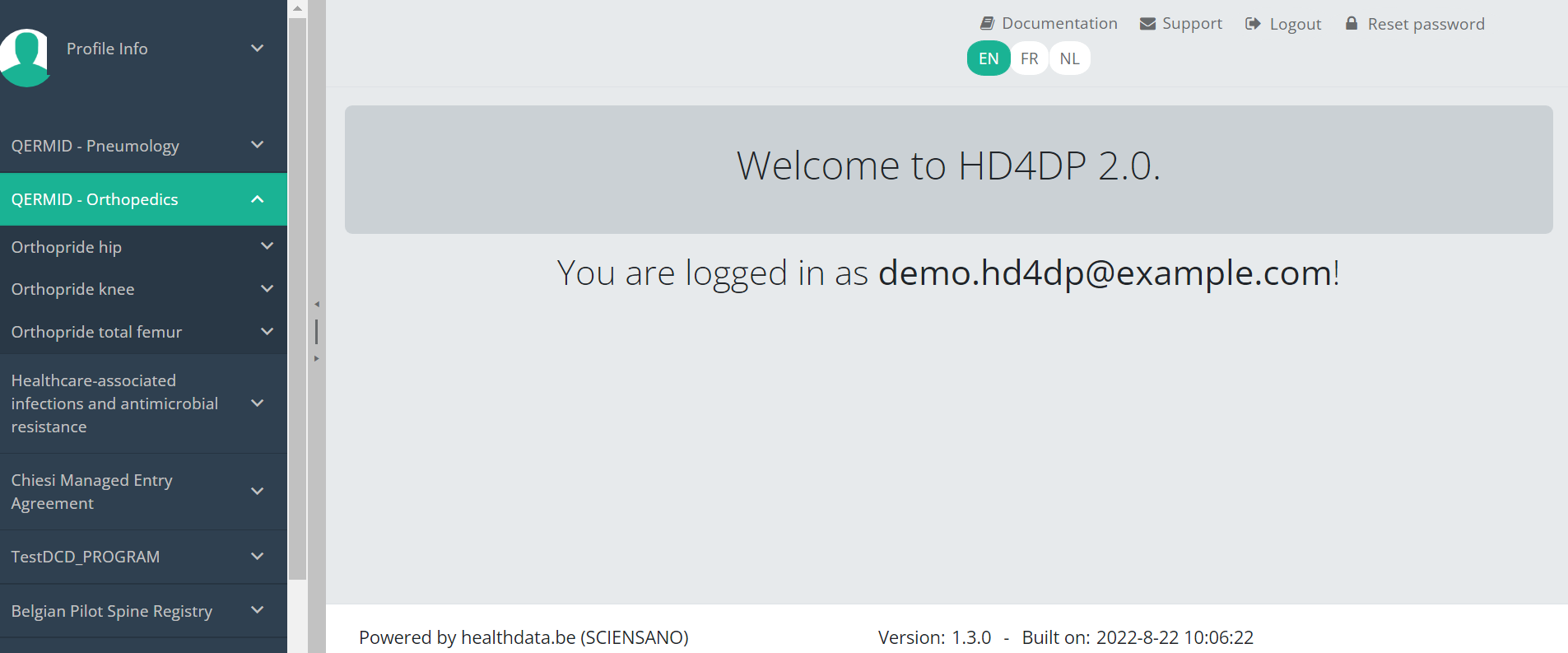
Select the study project Orthopride Hip.
You will see that the study project Orthopride Hip consists of three study sections: Primo-implantation, Revision and Resection.
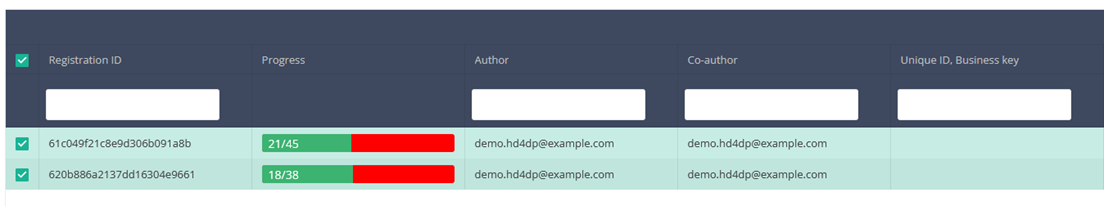
Create a registration
Create a registrationSuppose we want to create a "Primo-implantation" registration for the study project Orthopride Hip. In that case, we need to navigate to the study program QERMID Orthopedics and then to the study project Orthopride Hip. There we need to select "Primo-implantation" in the dark blue left menu.
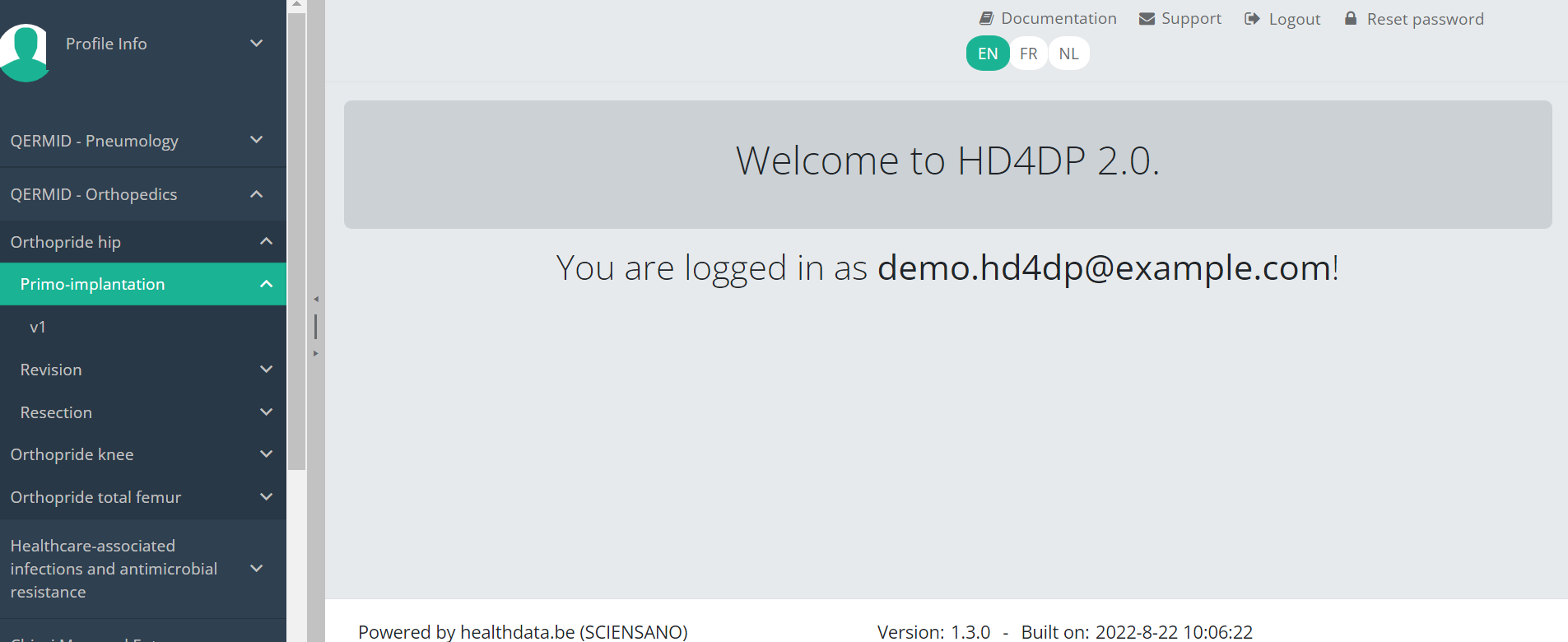
You will see the number of versions of this study section. In this case, there is only one version.
When you select the highest version of this study section for the first time, you will see an empty table in the main part of your screen. The table contains, among others, the following items: Registration ID, Progress, Author, Co-author, Unique ID, Business key, Registration code, National registry ID of the patient...
In the top right corner of the screen you can find a green button "+ New registration". Press this button.
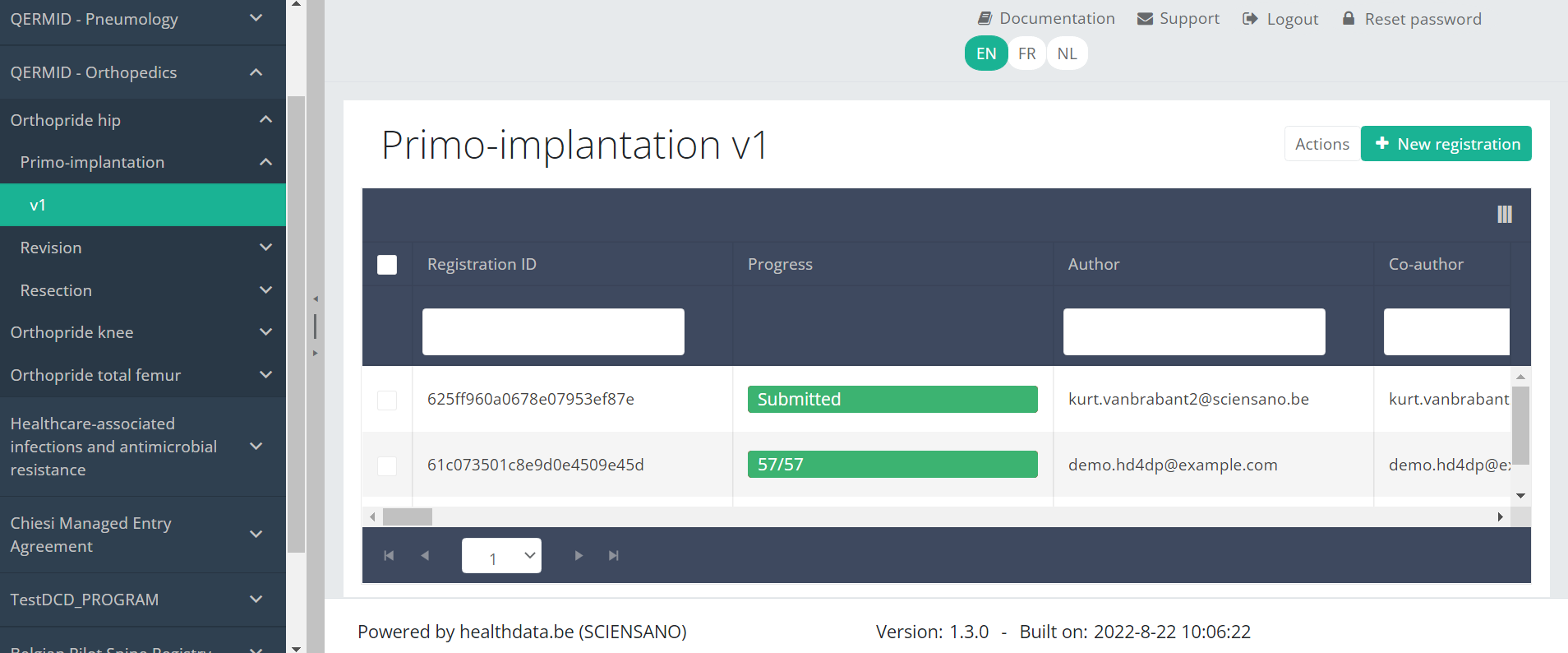
After pressing the button "+ New registration", the main screen will now be replaced with 2 sections: a study form (in the middle of the screen) and a Table of contents (on the right side of the screen).
By completing the study form, you will create a "Primo-implantation" registration for the study project Orthopride Hip.
The Table of contents indicates which sections you must complete. You can also use the table of contents to navigate through the study form: pressing a section in the table of contents will take you to this section in the study form.
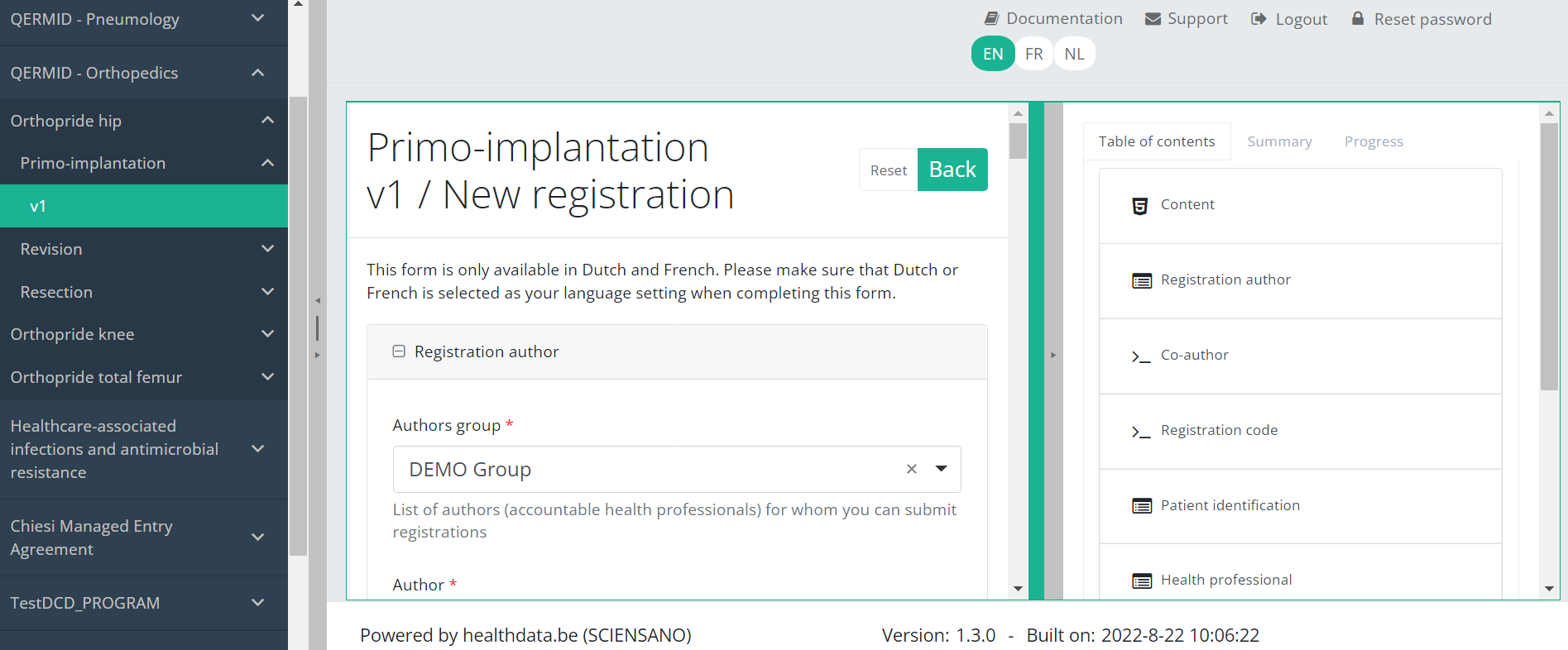
By pressing the tab "Progress" on the right side of the screen , the Table of contents will be replaced by a progress bar and a list of open validation errors.
You can use the list of open validation errors to navigate through the study form: pressing a validation error in the list will take you to this section in the study form.
When the study form is completed and there are no validation errors, you can Save or Submit this registration. Notice that the Submit button is in clear green.
When the study form is completed but there are validation errors, you can Save but not Submit this registration. Notice that the Submit button is in dim green.
When the study form is saved or submitted, the screen switches to the overview table. Now, this table is not empty anymore but shows the saved or submitted registration.
Find a registration
Find a registrationSuppose we want to find a "Primo-implantation" registration of the study project Orthopride Hip. In that case, we need to navigate to the study program QERMID Orthopedics and then to the study project Orthopride Hip. There we need to select "Primo-implantation" in the dark blue left menu.
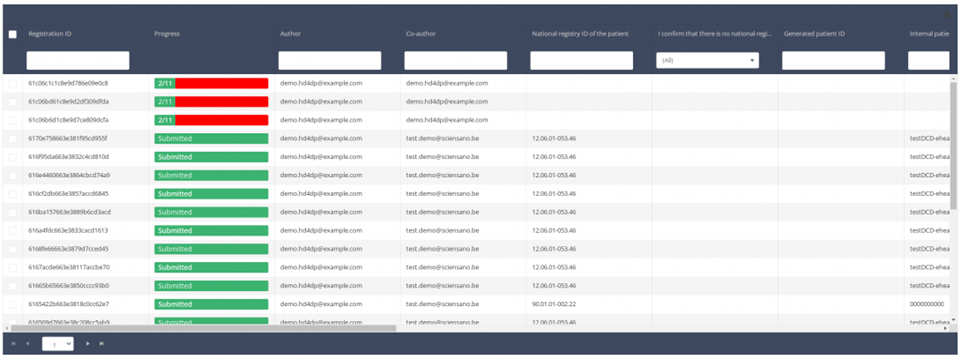
When you select a version of this study section, you will see the summary table in the main part of your screen. This table contains, among other things: Registration ID, Progress, Author, Co-author, Unique ID, Business Key, Registration Code, National Patient Registry Number…
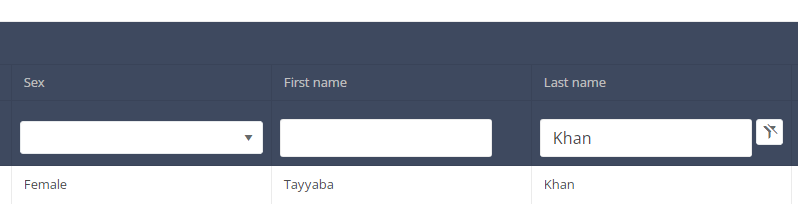
In the header of the summary table, you can use the filter below each column label. In the example below, the last name "Khan" has been entered in the filter (text field), so only the record with "Khan" is displayed.
Update a registration
Update a registrationSuppose we want to update a "Primo-implantation" registration of the study project Orthopride Hip. In that case, we need to navigate to the study program QERMID Orthopedics and then to the study project Orthopride Hip. There we need to select "Primo-implantation" in the dark blue left menu.
Important: a registration can be updated as long as the registration has not yet been submitted. If the status of a registration is "Saved" , the registration can still be updated.
When you select a version of this study, you will see the summary table in the main body of your screen. The table includes the following items: Registration ID, Progress, Author, Co-author, Unique ID, Business Key, Registration Code, National registry number of the patient.…
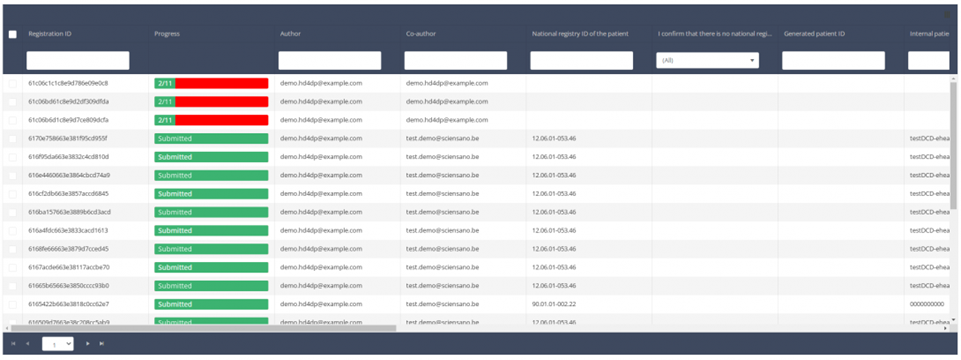
Use the filters in the header of the table to find the registration you want to update.
Once you have found the registration, you can open the registration's study form by clicking on the corresponding row in the summary table.
You can complete missing fields and/or change previously completed fields in the survey form.
At the end of the survey form, you can Save or Submit the registration.
A registration can be updated as long as it has the status "Saved" and as long as the registration not has been submitted. A submitted registration cannot be updated or deleted again.
Delete a registration
Delete a registrationSuppose we want to delete a "Primo-implantation" registration of the study project Orthopride Hip. In that case, we need to navigate to the study program QERMID Orthopedics and then to the study project Orthopride Hip. There we need to select "Primo-implantation" in the dark blue left menu.
Important: a registration can be deleted as long as the registration has not yet been submitted. If the status of a registration is "Open" , the registration can still be deleted.
When you select a version of this course of study, you will see the summary table in the main body of your screen. The table includes the following items: Registration ID, Progress, Author, Co-author, Unique ID, Business Key, Registration Code, National registry number of the patient...
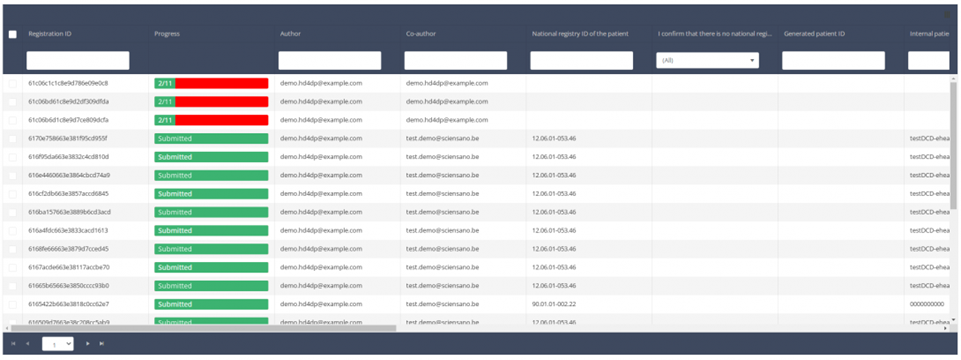
Use the filters in the header of the table to find the registration you want to delete.
Once you have found the registration you want to delete, you must select the registration by checking the checkbox at the beginning of the row in the summary table.
Then you need to press the "Actions" button at the top right of the summary table.
There are now two options, "Submit" and "Delete". Now press "Delete".
After you press "Delete," a pop-up message will appear asking you to confirm the deletion of the selected registration(s). If you are sure about this action, press "Confirm." If not, press "Cancel."
If you delete the registration, you cannot change its status or content.
The deleted registration will not be removed from the summary table. It remains present, but the status has changed from "Open" to "Deleted".
If you want to see only Open and Sent registrations, you can adjust the filter on the "Status" item in the summary table.
A registration can be deleted as long as the registration has not yet been submitted. If the status of a registration is "Open", the registration can still be deleted.
Submit a registration
Submit a registrationSuppose we want to submit a "Primo-implantation" registration of the study project Orthopride Hip. In that case, we need to navigate to the study program QERMID Orthopedics and then to the study project Orthopride Hip. There we need to select "Primo-implantation" in the dark blue left menu.
A registration can be submitted in two ways. The first way is at the end of the creation process using the study form (see: "Create a registration").
When the registration was completed using the study form, saved and there are no more validation errors, the registration can also be submitted via the overview table. This method can be useful to submit multiple registrations in the same action.
When you select a version of this course of study, you will see the summary table in the main body of your screen. The table includes the following items: Registration ID, Progress, Author, Co-author, Unique ID, Business Key, Registration Code, National registry number of the patient…
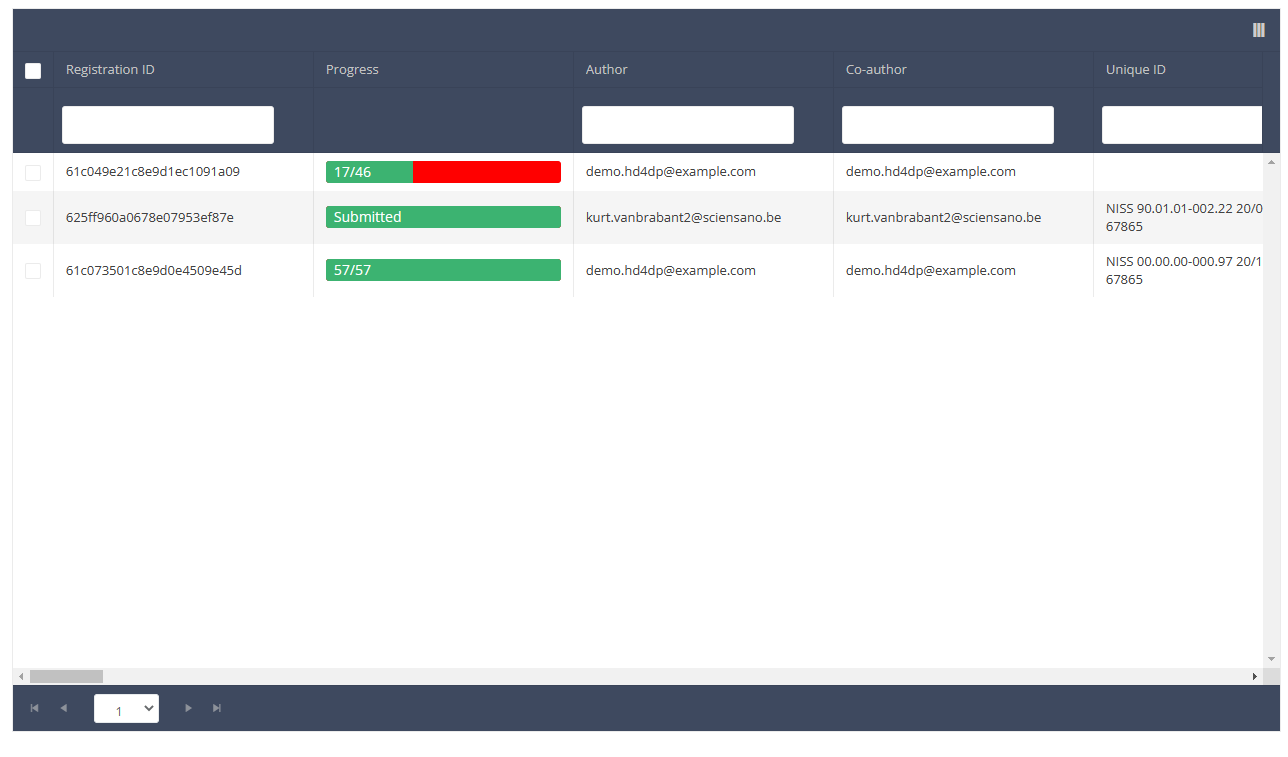
Use the filters in the header of the table to find the registration(s) you want to submit. For example, you can use the filters "Status" (set to "Open") and "Validation Errors" (set to "0") to find the registrations that are eligible for submission.
Once you have found the registration(s) you want to submit, you must select the registration(s) by checking the checkbox at the beginning of the row in the summary table.
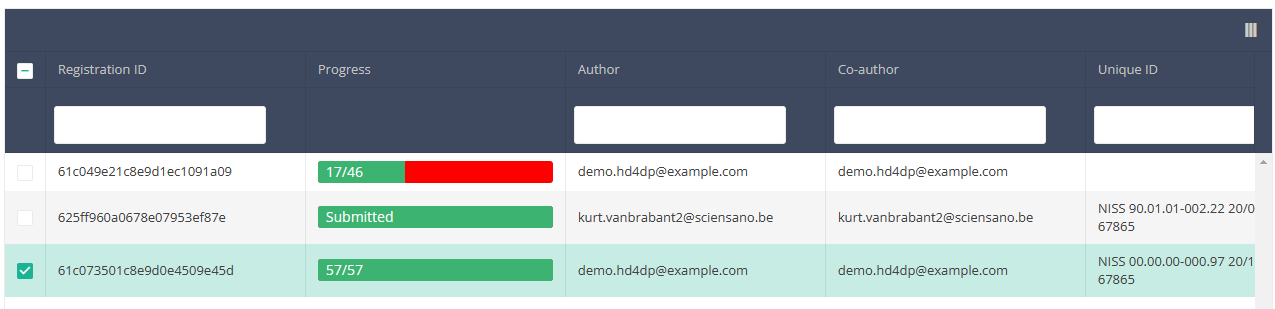
Then you need to press the "Actions" button at the top right of the summary table.
There are now two options, "Submit" and "Delete". Now press "Submit".
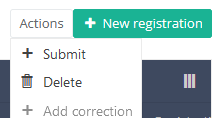
After you press "Submit," a pop-up message will appear asking you to confirm the submission of the selected registration(s). If you are sure about this action, press "Confirm." If not, press "Cancel."
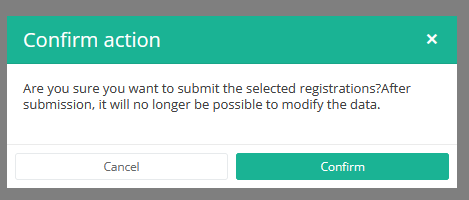
Once you confirm the submission, you cannot change the content of the registration(s). Sent registrations can also no longer be deleted.
The sent registration remains present in the summary table, but the status has changed from "Open" to "Submitted".
If you want to see only "Open" registrations, you can adjust the filter on the "Status" item in the summary table.
A registration can be submitted at the end of the creation process using the study form (see: Create a [project] registration).
When the registration was completed using the study form, saved and there are no more validation errors, the registration can also be submitted via the overview table. This method can be useful to submit multiple registrations in the same action.
Send a correction of a registration
Send a correction of a registrationTo send a correction of a submitted registration you need to submit the complete record again. An overview of the submitted corrections is available via the HD4DP v2 generic correction form.
The Add corrections function (and button) to a registration form has been discontinued. It is not available anymore, neither via the overview table, nor via the preview page of a registration.
Submitting the complete record again
The preferred way to send a correction is to fill out the complete registration form with the correct values and resubmit it. When you do so, the most recent version of the record that is received by healthdata.be will be considered to be the correct one.
Whether a record qualifies or not is determined by a so-called business key. This is a unique set of values of specific fields per record, such as a combination of the patient ID and the hospitalization date, or the niss code and the sample ID. The business key is created when submitting the original record, and so helps to identify the most recent record received in the healthdata.be database in case of resubmission.
Attention: In case one of the fields that build the business key needs to be corrected, the record that will be resubmitted will have a different business key. Consequently, both records will be considered as correct ones, since there is no identical business key.
You may consider to log a RITM ticket via our Service portal (https://sciensano.service-now.com/sp) to follow-up on this particular case.
There are two options to resubmit a record:
Option 1: Submission via S2S API / CSV Upload
The correction of the values is performed directly in the json or csv file. To resubmit the complete record via the back-end, please refer to the applicable technical documentation pages. The examples shown in the links below are for Pacemaker Primo-Implantation:
- For submitting the complete record as a .json file via S2S API, go here.
- For submitting the complete record as a .csv file via CSV Upload, go here.
Option 2: Submission via the HD4DP 2.0 web application

First, you need to access the HD4DP v2 web application, navigate to the study program and select the desired study project in the left dark blue menu. Then, fill out the complete registration form again manually with the correct values. Resubmit the completed registration form.
The previously submitted record will become irrelevant based on the business key.
The generic correction form
The Correction Form you can find in the list of study programmes and study projects in the HD4DP v2 web application contains an overview of all submitted corrections of registrations.
If you want to preview these corrections of registrations, navigate to the study program Correction form and then to the study project Correction form. Finally, select the most recent version.
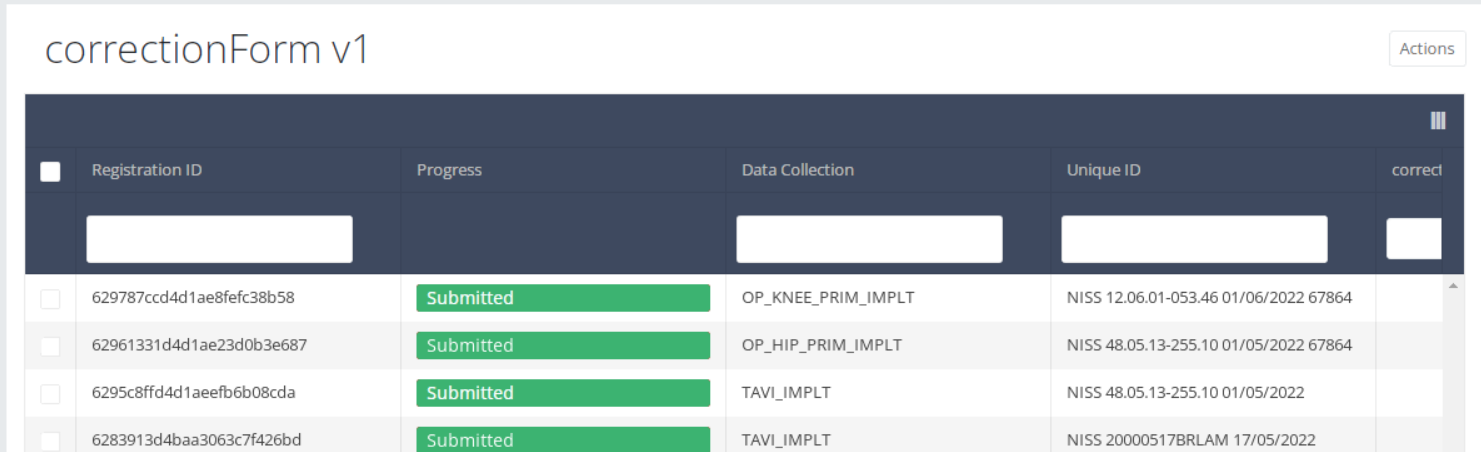
The corrections of registrations of the different projects will be displayed READ-ONLY in the overview table.
Registration statuses in HD4DP v2
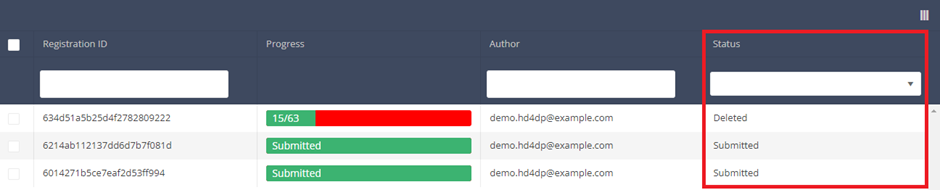
Registration statuses in HD4DP v2This article explains the different registration statuses in HD4DP v2.
Statuses are shown in Status column
You can select the columns you want to display via the menu Select visible columns located in the top-right corner:
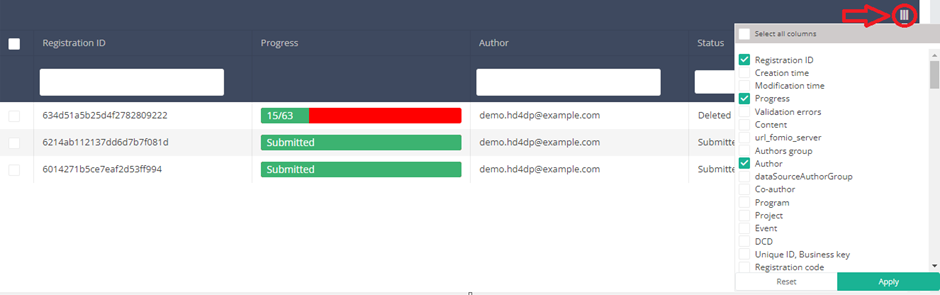
Select the columns you want to display and click on Apply.
Description of the statuses:
Open: Registration is created and stored. It has not been submitted
Deleted: Registration has been deleted.
Submitted: Registration has been submitted and sent.
Technical manual of the application HD4DP v2
Technical manual of the application HD4DP v2Technical documentation for IT services
HD4DP v2 Installation
HD4DP v2 InstallationHD4DP v2 Local is an application installed on the infrastructure of the Health Care Organisation participating in research projects facilitated by healthdata.be.
The installation of HD4DP v2 Local is executed by the DevOps team of healthdata.be.
Server Installation and Configuration
Installing and configuring the server requires the following actions:
- install and configure the server as described in the article "HD4DP 2.0 Infrastructure instructions";
- complete and return to support.healthdata@sciensano.be the server configuration file "HD4DP v2 Infrastructure Sheet".
The HD4DP v2 application is more modular and will support scaling up to meet the requirements of the various data collection projects we facilitate. It will offer several micro-services that will run concurrently on the same machine.
The server should therefore require more resources than the one currently hosting the HD4DP 1.0 application. Furthermore, the resources allocated should be increased. It is therefore on the one hand imperative to use virtualization for the creation of the machine. On the other hand. It is also imperative to store files and make regular backups to a file server.
Below we take up our three categories of organizations sending data to healthdata.be and the resources we recommend allocating to their virtual machine:
- "Small": Small data provider;
- "Medium": Medium data provider;
- "Large": Big data provider.
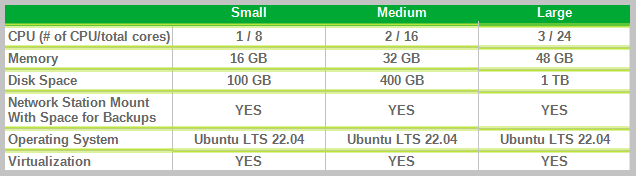
Finally, we also offer the possibility for each hospital to have an integration server and a production server. Healthdata.be will deploy the new release of the application on the integration server. This will allow you to accept or decline the promotion of a new release of the HD4DP 2.0 application to the production server. This option is highly recommended, but not mandatory.
Therefore, could you answer the question: Do you want to first deploy HD4DP on an integration server? Yes/No. If Yes, Could you provide a server whose label used for specifications is "Small" (following the instructions in section 1 of this mail), that is:
- Processors number: 1
- Physical cores/Processor: 8
- RAM memory: 16 Go
- Disk space: 100 Go
- Network Station Mount with Space for Backups
- Operating System: Ubuntu LTS v22.04
- Virtualization
Server installation timing
In order to establish the deployment schedule for the HD4DP 2.0 application within your organization, we would like to know when the server could be installed and configured. To this end, could you give us the 2 dates relating to the installation of the server:
- Starting date;
- Finalization date.
Based on these dates, an employee of healthdata.be will regularly monitor the operations linked to the installation of the server.
For any request for information on installing the HD4DP 2.0 server, please send an email to hd-architecture-20@sciensano.be.
HD4DP v2 Infrastructure instructions
HD4DP v2 Infrastructure instructionsIntroduction
This document is written for IT staff / system engineers of data providers and therefore assumes technical knowledge. It acts as a guide through the on-boarding process of HD4DP v2 and covers installation of the server, user configuration, network configuration and remote access.
The order of steps in this document should be respected during execution.
If you need more informationor have any questions, feel free to contact us.
Overview
HD4DP v2 consists of a modular application stack, which allows Healthdata to seamlessly upgrade individual elements.
An HD4DP v2 deployment comprises of following components:
- Form.io component
- MongoDB
- PostgreSQL
- Nextgen Connect
As it is the case in HD4DP 1.0, an Encryption Module with a connection to the eHealthBox is still required for HD4DP v2 and must be provided by the data provider.
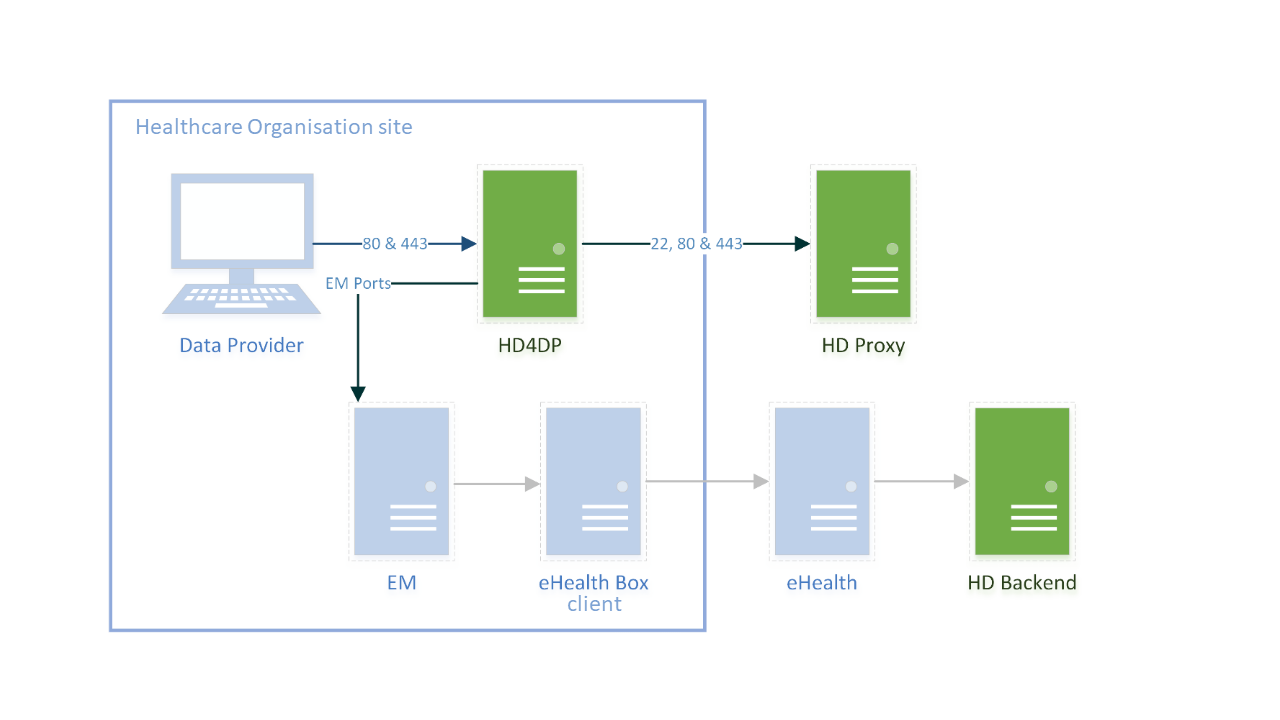
Network configuration
IP
The HD4DP server needs to be accessible via domain names in DNS, and must have a static IP in your private network.
DNS
The application stack of HD4DP v2 requires four domain names pointing to the IP of the locally installed HD4DP v2 server. Use the following names in your DNS:
- nextgenconnect.hd4dp.<yourdomain.be>
- hd4dp.<yourdomain.be>
- metabase.hd4dp.<yourdomain.be>
- admin.hd4dp.<yourdomain.be>
Firewall
The following connections should be possible in the firewall flow:
- To and from (a) machine(s) in your IT department on port 22 for initial configuration and local support.
- To and from the Encryption Module server. The protocol and ports depend on your local EM implementation. Contact your EM vendor if more information is necessary.
- Reachable by your staff who uses HD4DP, on ports 80 and 443 for HTTP(s) traffic.
- To and from the LDAP server (this is not mandatory if you are not using LDAP to authenticate) (port 389 by default)
The Healthdata proxy server is used as a gateway to the internet for the security of HD4DP servers. The configuration of this proxy server will be provided to you by Healthdata at a later date.
Server installation
To install the application stack of HD4DP v2, Healthdata requires a fresh installed operating system, specifically Ubuntu Server 22.04 LTS.
Please use these instructions even if you have previous experience with installing this operating system, as its configuration is specific for Healthdata.
These instructions assume that the network configuration described in the previous section is completed.
Instructions
HD4DP v2 requires a (virtual) machine running Ubuntu Server 22.04 LTS.
We assume knowledge of loading an .iso file onto a (virtual) machine. Healthdata can’t provide instructions for this, as the environment of your center is unknown. Should you have any trouble, however, please contact Healthdata support so that we can help out.
Please find the installation steps below.
Installation steps
- Download the .iso file from the link below.
Download Ubuntu Server 22.04 LTS - Create a new (virtual) machine with Linux Ubuntu 64 bit as the OS family
- When prompted, select the .iso file downloaded in step 1.
- After some time, you will be prompted to select a system language. Select English.
- “Keyboard configuration”
Select your preferred keyboard layout and press enter - “Network Connections”
Highlight the network interface and press enter. Navigate as follows:
Edit IPv4 -> Manual -> enter the network details -> save -> Done - Proxy IP -> Leave default/empty.
- “Configure Ubuntu Archive Mirror” -> leave default
- “File system Setup” -> Use An Entire Disk
- Proceed until “Confirm destructive action” -> press continue. The installation process starts, this can take several minutes.
- In the meantime, create the user for Healthdata.
username = healthdata,
Password = choose a secure password and communicate it to Healthdata. - Mark “Install OpenSSH server”. This will be used for remote access. “Import SSH Identity” -> no -> done
- “Featured Server Snaps” -> Select nothing and press Done.
- Wait until installation is finished.
Configuration steps
Connecting to the server
Log into the machine with the Healthdata user created in the previous section.
Instructions (from a Windows machine):
- Install the tool Putty and open the application.
- On the configuration screen, enter the following (replace cursive text with the appropriate values)
- Host Name: healthdata@server_private_ip
- Port: 22
- Connection type: SSH
- Click Open. Enter the Healthdata password (you will not see text as you type, you can paste into putty by right-clicking in the terminal).
- You should now be logged in and see a prompt “healthdata@server_name:~$”
Administrator account for internal use
An administrator account for internal use can be created on the HD4DP v2 server.
The configuration of remote access (described below) should not happen on this account, but on the Healthdata account.
The internal account can later be used to install and configure OS monitoring software and antivirus software by the internal IT team. For more information, see the section on Antivirus and Monitoring.
(Text with a gray background should be entered as a command in the terminal of the server)
Create the user:
sudo adduser <username>
Add the user to the sudo group
sudo usermod -aG sudo <username>
Installation and configuration of the software stack
Healthdata.be support will instruct you when to execute the next step, which is to enable remote access so that Healthdata can execute the software installation and configuration.
Backups
The configuration of the HD4DP v2 server is administered by Healthdata and does not require backups.
HD4DP v2 regularly dumps its databases automatically to the /backup directory on the server. A network storage should be mounted at this location.
Please fill out the infrastructure sheet with the required credentials, domain name/url, protocol… to connect to the network drive. The connection will then be configured by Healthdata.
Patching and Updates
Healthdata configures HD4DP v2 servers to automatically receive recommended security updates. The choice for Ubuntu 22.04 is motivated by the long-term support for this version. Security flaws are rare in this distribution, and security updates are quick and often don’t require a system reboot.
If the IT department of your organization prefers to manage patches, this is possible but not encouraged. Please use the account for internal use created in Section Server installation for this purpose.
Antivirus and Monitoring
Most data providers will want to manage their own antivirus and OS monitoring on all machines in their network. Installation of such software on the HD4DP v2 server is allowed, but Healthdata should be informed about all extra software installed on the server. Additionally, Healthdata will not provide support for the installation of this software.
Contact information
Email: support.healthdata@sciensano.be
Phone: +32 2 793 01 42
HD4DP v2 Infrastructure Sheet
HD4DP v2 Infrastructure SheetThe HD4DP v2 Infrastructure Sheet contains information that healthdata.be needs in order to start the installation of the HD4DP 2.0 Software at your organization.
Below you can find the description of the necessary information:
SERVER CONNECTION
Healthdata.be performs its installation and support tasks remotely (using VPN or remote port forwarding via SSH). Please provide the required credentials.
- Type of connection (VPN / Remote port forwarding via SSH)
- Link (IF VPN)
- Username, token, other (if VPN)
- Password (if VPN)³
- Public SSH Key (if remote port forwarding)
³ For security reasons, we advise to communicate passwords to us either by phone, or via a link using a secret-sharing service such as onetimesecret.com.
SERVER MACHINE
- Server Name
- Internal IP-Address
- Ram (in GB)
- CPU (number of CPU's and number of cores)
- Disk space (in GB)
- Username: Healthdata
- Password ³
³ For security reasons, we advise to communicate passwords to us either by phone, or via a link using a secret-sharing service such as onetimesecret.com.
ATTACHED DRIVE FOR BACKUPS
HD4DP 2.0 regularly performs data dumps for backup purposes. Please provide connection information to a network share volume.
- Link / IP address
- Path
- Username
- Password ³
³ For security reasons, we advise to communicate passwords to us either by phone, or via a link using a secret-sharing service such as onetimesecret.com.
USER MANAGEMENT
HD4DP can either connect to a LDAP server or use its own application database for performing user authentication and management. Please check the user management mechanism you want to use.
- LDAP user management : Yes / No
- Application user management : Yes / No
LDAP configuration (Optional)
If you chose ‘LDAP user management’ as user management mechanism, please provide the following information that allows us to connect to your LDAP system.
- Connection URL
- Username
- Password³
³ For security reasons, we advise to communicate passwords to us either by phone, or via a link using a secret-sharing service such as onetimesecret.com.
SOFTWARE CONFIGURATION
Encryption Module interface
HD4DP communicates with the Encryption Module (EM) either using the file system interface or by calling a REST web service. Please choose which interface HD4DP should use for its communication with the Encryption Module.
- REST web service
- File system
REST web service interface
If you chose to communicate with the Encryption Module using a REST interface, please provide the web service URLs that should be used by HD4DP for its communication with EM.
- "Outgoing flow URL: Example: http://host:8080/encryptionmodule/send"
- "Incoming flow URL : Example: http://host:8080/encryptionmodule/receive"
File system interface
- "Incoming directory: Directory where HD4DP checks for incoming files"
- "Incoming directory: Directory where HD4DP writes outgoing files"
- "Incoming directory: Directory to which HD4DP moves successfully processed files"
- "Incoming directory: Directory to which HD4DP moves unsuccessfully processed files"
Requirements for the HD4DP installation
Requirements for the HD4DP installationThis documentation details the necessary adaptations to be performed in order to allow the necessary technical accesses and smooth operation of the different healthdata.be platforms and interfaces.
The file is available for download below.
HD4DP v2 S2S API
HD4DP v2 S2S APIIntroduction
The HD4DP v2 S2S API is a unified Application Programming Interface (API) that will allow participating Healthcare Organizations (HCO) to submit DCDs data to HD4DP2.0 fully automated.
In the manual of the application HD4DP v2 we provide detailed information about the S2S API. Please read this documentation before its project specific use.
- End-to-End process to submit DCD registrations
- API Endpoint for supporting the DCD submit process
- MDM Field description - DB Model
- Swagger API
- Postman collection
Important note:
For code fields (fieldType = 'CODE') the id of the codeListValue item must be sent, not the code value or the label. In future releases it will be made possible to also send the code value.
Training
Below you can review the S2S API training organized by healthdata.be:
1. End-to-End process to submit DCD registrations
1. End-to-End process to submit DCD registrationsTable of contents
- API description
- Get organizations
- Get the DCD menu structure
- Get DCD field definitions
- Get DCD payload example
- Submit DCD registrations
- User password
- Submission of drafts
- Adding separators to a NISS number
API description
The baseURL to be used has to following built: https://<hd4dp_url>/proxy
| API end-point | Response | Authentication | Notes |
|---|---|---|---|
| {{baseURL}}/api/organizations | List of organizations. Client must select the right organizationId | No | Current existing end-point is: /api/installation/organizations We’ll create this new end-point with a different signature re-routing the call to this existing one or we will refactor the existing one to this new signature. |
| {{baseURL}}/api/dcd/menu/structure? organization-id={organizationId} | List of projects of the given organization, dcds of each project, dcdVersions of each dcd in a JSON format Client can get dcdId and dcdVersionId (optional) which are needed on following API calls. | No | https://github.com/Sciensano-Healthdata/hd4-formio/blob/develop/dev/mock_menu_structure/menu-structure.json |
| {{baseURL}}/api/dcd/payload/definition? dcd-id={dcdId}; <optional>version={version}; <optional>language-id={languageId} | List of all the fields of the form as well as their corresponding data-types that are allowed in the json data structure for the Payload | Yes | This field names values are the key properties in the formIO json config form. When we implement this new api end-point, we need to parse the json content in order to get the key properties. Given these field keys, we’ll get each field definition from new API end-points helpers: /api/dcd/field?field-id={fieldId} /api/dcd/codelist?codelist-id={codelistId} These ones are described in the next table. <optional parameter> version={version} : If this parameter is not provided, latest one is assumed <optional parameter> language-id = {languageId} : language id for the code_list example results. If this parameter is not provided, default language will be English. Current permitted values: en: English nl: Dutch fr: French Client must build this json object as the payload data to be sent based on this list of fields, on the last api call |
| {{baseURL}}/api/dcd/payload/example?dcd-id={dcdId}; <optional>version={version} | Example of payload in JSON format | Yes | Providing this API end-point in order to help the Client on the Payload build with an example <optional parameter> version={version} : If this parameter is not provided, latest one is assumed |
| {{baseURL}}/api/dcd/payload/submit? organization-id={organizationId}; dcd-id={dcdId}; <optional>version={version}; <optional>data-src-type={dataSrcType}; POST Payload | Results info if succeed Error info if failed | Yes | Some implementation tasks is needed in here in order to return the result info (either succeed or failed). Similar like the one in HDConnectProxyRestTemplate.postCsv method, and the CsvExecutionResult object build. <optional parameter> version={version} : If this parameter is not provided, latest one is assumed. <optional parameter> data-src-type={dataSrcType} : permitted values: API CSV If this parameter is not provided, default values is <HD4DP>. |
| {{baseURL}}/api/dcd/payload/submit?organization-id={organizationtId};dcd-id={dcd-id};<optional>dcd-version-id={dcdVersionId};<optional>incl-submit-data={inclSubmitData}; <optional>incl-submit-results={inclSubmitResults}; GET method | List of submitted dcds data and/or their corresponding business keys or validation errors. | Yes | <optional parameter> dcd-version-id={dcdVersionId} : If this parameter is not provided, lastest one is assumed <optional parameter> incl-submit-data={inclSubmitData} : If this parameter is not provided, default values is <false> <optional parameter> incl-submit-results={inclSubmitResults} : If this parameter is not provided, default values is <true> |
Get organizations
Request example
GET {{baseURL}}/api/organizations
Request response
[
{
"organizationId": 1,
"organizationCode":"12345",
"name":"First Organization Name",
"loginMethods":[
"USERNAME_PASSWORD",
"EID"
]
},
{
"organizationId": 2,
"organizationCode":"32154",
"name":"Second Organization Name",
"loginMethods":[
"EID"
]
}
]
Get the DCD menu structure
Request example
GET {{baseURL}}/api/dcd/menu/structure?organization-id={organizationId}
Request parameters
- {organizationtId}: Id of the organization for which we want to list the menu structure
- {{baseURL}}: the value for this field is https://<hd4dp_url>/proxy
Request response
All the menu structure for that organization, including projects, dcds of each project and dcd versions of each dcd, in a JSON format.
[
{
"id": 4,
"key": "zephyr_pneumo_program",
"projects": [
{
"id": 4,
"key": "zephyr_pneumo_project",
"dcds": [{
"id": 9,
"key": "zephyr_pneumo_t1_dcd",
"dcdVersions": [{
"id": 9,
"version": 1,
"supportsEN": false,
"supportsFR": true,
"supportsNL": true
}]
},
{
"id": 10,
"key": "zephyr_pneumo_tr_dcd",
"dcdVersions": [{
"id": 10,
"version": 1,
"supportsEN": false,
"supportsFR": true,
"supportsNL": true
}]
},
. . .
. . .
. . .
. . .
]
Get DCD field definitions
Request example
GET {{baseURL}}/api/dcd/payload/definition?dcd-id={dcdId};version={version};language-id={languagerId}
Request parameters
- {dcdId}: Id of the dcd
- <optional parameter> {version}: If this parameter is not provided, latest one is assumed
- <optional parameter> {languageId}: language id for the code_list example results. If this parameter is not provided, default language will be English. Current permitted values:
- en: English
- nl: Dutch
- fr: French
Request response
{
"CD_SURGL_APPR_FEMO": {
"field_type": "CODE",
"data_type": "number",
"code_list": [
{
"ID": 68224,
"CODE_VALUE": "870646003",
"LABEL_EN": "Femoral (Hemi)"
},
{
"ID": 68225,
"CODE_VALUE": "465954006",
"LABEL_EN": "Femoral + Cup"
}
]
},
"D_IMPLANT": {
"field_type": "DATE",
"data_type": "timestamp",
"code_list": null
},
"TX_TPE_INSTRU": {
"field_type": "FREE TEXT",
"data_type": "string",
"code_list": null
},
"MS_PAT_HGHT": {
"field_type": "FREE TEXT",
"data_type": "number",
"code_list": null
}
}
Get DCD payload example
Request example
GET {{baseURL}}/api/dcd/payload/example?dcd-id={dcdId};version={version}
Request parameters
- {dcdId}: Id of the dcd
- <optional parameter> {version}: If this parameter is not provided, latest one is assumed
Request response
Given the previous Payload field definition example on previous chapter, we build a payload content example accordingly.
{
"CD_SURGL_APPR_FEMO": 68224,
"D_IMPLANT": "2021-04-30T22:00:00",
"TX_TPE_INSTRU": "P-432",
"MS_PAT_HGHT": 180
}
Submit DCD registrations
Request example
POST {{baseURL}}/api/dcd/payload/submit?organization-id={organizationId};dcd-id={dcdId};version={version};data-src-type={dataSrcType}
Header:
MediaType.APPLICATION_JSON
Body:
{
{
"CD_SURGL_APPR_FEMO": 68224,
"D_IMPLANT": "2021-04-30T22:00:00",
"TX_TPE_INSTRU": "P-432",
"MS_PAT_HGHT": 180
},
{
"CD_SURGL_APPR_FEMO": 68225,
"D_IMPLANT": "2021-04-14T22:00:00",
"TX_TPE_INSTRU": "P-545",
"MS_PAT_HGHT": 1209
},
{
"CD_SURGL_APPR_FEMO": 68224,
"D_IMPLANT": "2021-05-01T22:00:00",
"TX_TPE_INSTRU": "T-678",
"MS_PAT_HGHT": 210
}
}
Request parameters
- {organizationtId}: Id of the organization for which we want to submit the DCD registration.
- {dcdId}: Id of the DCD we want to submit.
- <optional parameter> {version}: Id of the DCD Version we want to submit. If this parameter is not provided, latest one is assumed.
- <optional parameter> {dataSrcType}: The data source type e.g: API or CSV.
Request payload
- {Header}: MediaType.APPLICATION_JSON
- {Body}: JSON object with an array of DCDs data to be submitted, following the specifications and examples provided by the described api end-points:
- GET /api/dcd/payload/definition
- GET /api/dcd/payload/example
Request response
Client get a response per each DCD line. If the DCD submission was successful, Client will get a TXT_BUSINESS_KEY value. If it was failed, Client will get an error detailed info: Http Status Code, Name and Exception details:
{
"TX_BUSINESS_KEY": "NISS 12.06.01-052.46 30/04/2021 67864",
{
"HTTP_STATUS_CODE": 405,
"HTTP_STATUS_NAME": "Method Not Allowed",
"HTTP_STATUS_EXCEPTION_DETAILS": "Exception details for Method Not Allowed example",
},
"TX_BUSINESS_KEY": "NISS 12.06.01-071.48 01/05/2021 67864",
}
User / password
The username and password can be requested at our Servicedesk.
Submission of drafts
To submit a draft, the only thing to add is the key STATUS (all upper case) with the value "draft" to the request, like this:
"STATUS": "draft",
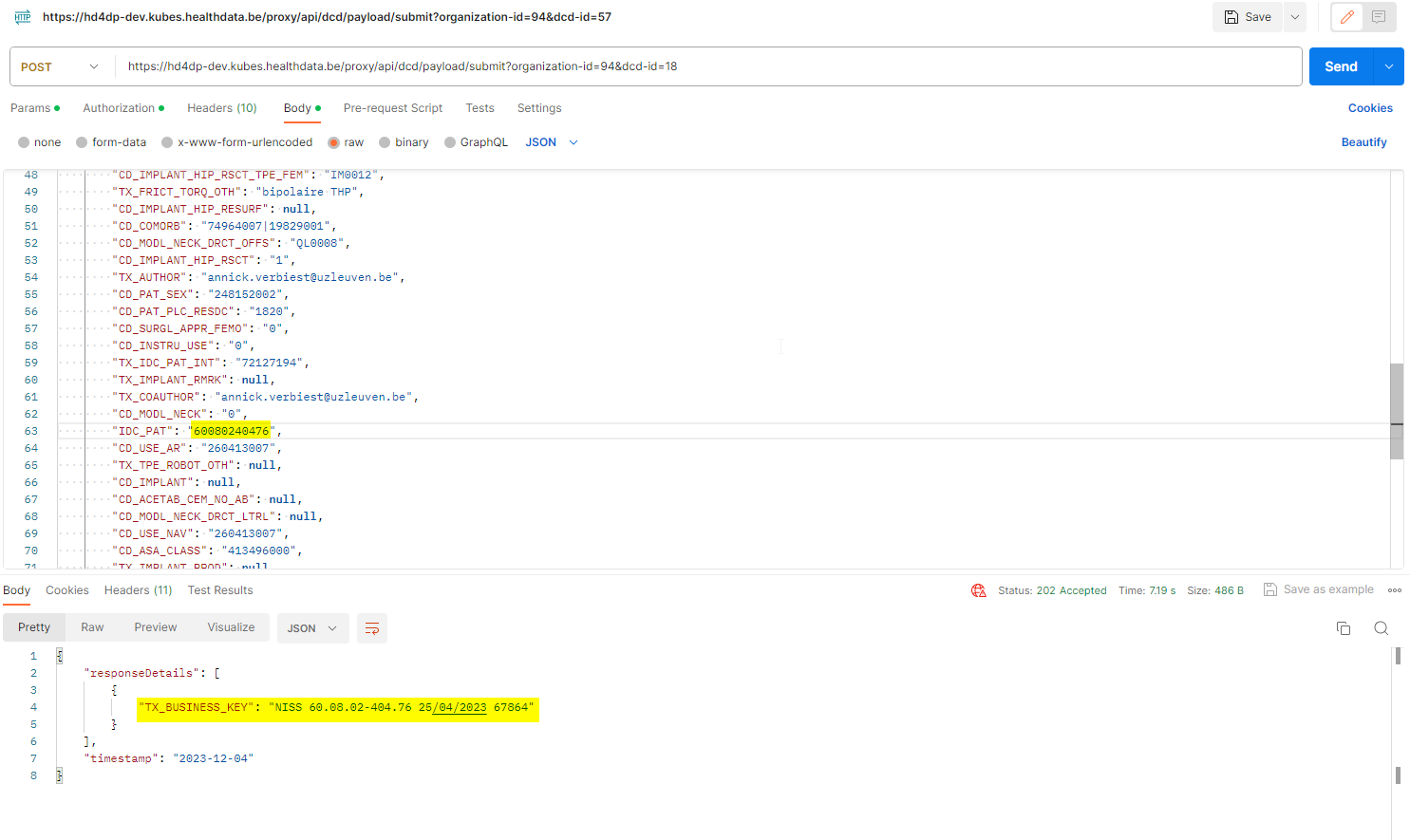
If the status is sent as "draft", a draft registration is created. In case of a successful submission the user receives a HTTP 202 (Accepted) notification with an empty business key, like this:

If the status key is not sent, the default value "submit" is used. The user performs a complete submission, and in case of a successful submission the user receives a HTTP 202 (Accepted) notification with a business key, like this:

Note that only "draft" status needs to be sent when sending a draft. The status "submit" is not necessary when performing a complete submission.
In the GUI, the difference is that the draft submission has the status open while the complete submission has the status submitted:
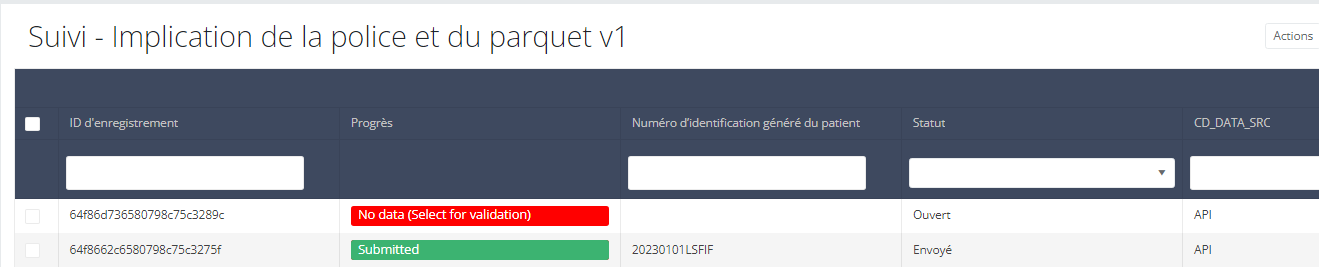
Once the draft is submitted in the GUI, the business key is generated and the process completed.
Adding separators to a NISS number
It is not necessary to add separators in a NISS number when uploading a file using S2S API. You can fill the NISS number out both with or without seperators. E.g.: 85.04.02-169.32 or 85040216932.
2. API Endpoint for supporting the DCD submit process
2. API Endpoint for supporting the DCD submit processTable of contents
- API description
- Get DCD field definition list
- Get DCD code list values
- Get DCD menu translations
- Get DCD field definition translations
- Search for SADMI medical devices
- Adding repeatable blocks to payload
API description
The baseURL to be used has to following built: https://<hd4dp_url>/proxy
| API end-point | Response | Notes |
|---|---|---|
| {{baseURL}}api/dcd/field? dcd-id={dcdId}; <optional>dcd-version-id={dcdVersionId}; <optional>included-codelist-values={inclCodelistValues}; <optional>field-name={fieldname}; <optional>language-id={languageId} | List of DCD fields definition, even with codelist values if the field is CODE field type. | <optional parameter> dcd-version-id={dcdVersionId} : If this parameter is not provided, latest one is assumed <optional parameter> included-codelist-values={inclCodelistValues}: permitted values: true False If this parameter is not provided, default value is <true>. <optional parameter> field-name={fieldname}: A field name e.g: TX_AUTHOR_GR <optional parameter> {languageId} : language id for the code_list example results. If this parameter is not provided, default language will be English. Current permitted values: en: English nl: Dutch fr: French |
| {{baseURL}}/api/dcd/codelist? code-list-id={codelistId}; <optional>language={language} | List of values for the codelist of the given codelistId.(List<CodeListItem>) | Checking and getting this info based on MDM -> CODE_LIST table and the relationed tables (CODE, CODE_CONTENT, etc.) See MDM – Field description – DB model diagram at the Attachment chapter for more details. <optional parameter> {languageId} : language id for the code_list example results. If this parameter is not provided, default language will be English. Current permitted values: en: English nl: Dutch fr: French |
| {{baseURL}}/api/dcd/menu-translations? dcd-id={dcdId}; <optional>dcd-version-id={dcdVersionId}; <optional>language-id={languageId} | <optional parameter> dcd-version-id={dcdVersionId} : If this parameter is not provided, lastest one is assumed <optional parameter> {languageId} : language id for the code_list example results. If this parameter is not provided, default language will be English. Current permitted values: en: English nl: Dutch fr: French | |
| {{baseURL}}/api/dcd/field/translation? dcd-id={dcdId}; <optional>dcd-version-id={dcdVersionId}; <optional>language-id={languageId}; <optional>field-name={fieldName} | List of fields translations for a given dcdId in the specified languageId (English if it is not provided) in a JSON format. Optional fieldId parameter can provided just for getting this info but only for this fieldId. | <optional parameter> dcd-version-id={dcdVersionId} : If this parameter is not provided, lastest one is assumed <optional parameter> {languageId} : language id for the code_list example results. If this parameter is not provided, default language will be English. Current permitted values: en: English nl: Dutch fr: French <optional parameter> field-name={fieldName}: A field name e.g: TX_AUTHOR_GR |
| {{baseURL}}/api/externalreference/sadmi? notificationCode={notificationCode}; name={name}; reference={reference}; distributorName={distributorName}; manufacturerName={manufacturerName}; classificationCode={classificationCode} | List of Medical Device Types: NotificationCode Name Reference URL Distributor Manufacturer Classification State Client must select the right info needed. |
Get DCD field definition list
Request example
GET {{baseURL}}/api/dcd/field?dcd-id={dcdId};dcd-version-id={dcdVersionId};included-codelist-values={inclCodelistValues};field-name={fieldName};language-id={languageId};
Request parameters
- {dcdId} : Id of the menu dcd which we want to get all the translations.
- <optional parameter> {dcdVersionId} : If this parameter is not provided, latest one is assumed.
- <optional parameter> {includedCodelistValues}: boolean value (true or false). If this parameter is not provided, default value will be true. If true and for those fields with type equal to CODE, codelist values will be provided in the result.
- <optional parameter> {fieldName}: A field name e.g: TX_AUTHOR_GR
- <optional parameter> {languageId} : language id for the menu options example results. If this parameter is not provided, default language will be English. Current permitted values:
- en: English
- nl: Dutch
- fr: French
Request response
with parameter includeCodelistValues equal to true (default value)
{
"TX_TPE_INSTRU": {
"field_type": "FREE TEXT",
"code_list": null
},
"D_IMPLANT": {
"field_type": "DATE",
"code_list": null
},
"MS_PAT_XXXX": {
"field_type": "UNKNOWN",
"code_list": null
},
"MS_PAT_HGHT": {
"field_type": "CODE",
"code_list": [
{
"ID": 68224,
"CODE_VALUE": "870646003",
"LABEL_EN": "Femoral (Hemi)"
},
{
"ID": 68225,
"CODE_VALUE": "465954006",
"LABEL_EN": "Femoral + Cup"
}
]
}
}
with parameter includeCodelistValues equal to false
{
"TX_TPE_INSTRU": {
"field_type": "FREE TEXT",
"code_list": null
},
"D_IMPLANT": {
"field_type": "DATE",
"code_list": null
},
"MS_PAT_XXXX": {
"field_type": "UNKNOWN",
"code_list": null
},
"MS_PAT_HGHT": {
"field_type": "CODE",
"code_list": 23
}
}
Get DCD code list values
Request example
GET {{baseURL}}/api/dcd/codelist?codelist-id={codelistId};language-id={languageId}
Request parameters
- {codelistId} : Id of codelist which we want to get the permitted values
- <optional parameter> {languageId} : language id for the code_list example results. If this parameter is not provided, default language will be English. Current permitted values:
- en: English
- nl: Dutch
- fr: French
Request response
{
{
"ID": 68224,
"CODE_VALUE": "870646003",
"LABEL_EN": "Femoral (Hemi)"
},
{
"ID": 68225,
"CODE_VALUE": "465954006",
"LABEL_EN": "Femoral + Cup"
}
}
Get DCD menu translations
Request example
GET {{baseURL}}/api/dcd/menu-translations?dcd-id={dcdId};dcd-version-id={dcdVersionId};language-id={languageId}
Request parameters
- {dcdId} : Id of the menu dcd which we want to get all the translations.
- <optional parameter> {dcdVersionId} : If this parameter is not provided, lastest one is assumed.
- <optional parameter> {languageId} : language id for the menu options example results. If this parameter is not provided, default language will be English. Current permitted values:
- en: English
- nl: Dutch
- fr: French
Request response
Example 1: languageId = “en” - English language menu translations
{
{
"zephyr_ortho_program": "QERMID - Orthopedics",
"zephyr_ortho_knee_project": "Orthopride knee",
"zephyr_ortho_knee_t1_dcd": "Primo-implantation"
},
{
"zephyr_ortho_program": "QERMID - Orthopedics",
"zephyr_ortho_knee_project": "Orthopride knee",
"zephyr_ortho_knee_t2_dcd": "Revision"
}
. . . . .
}
Example 2: languageId = “nl” - Dutch language menu translations
{
{
"zephyr_ortho_program": "QERMID - Orthopedie",
"zephyr_ortho_knee_project": "Orthopride knie",
"zephyr_ortho_knee_t1_dcd": "Primo-implantatie"
},
{
"zephyr_ortho_program": "QERMID - Orthopedie",
"zephyr_ortho_knee_project": "Orthopride knie",
"zephyr_ortho_knee_t2_dcd": "Revisie"
},
. . . . .
}
Example 3: languageId = “fr” French language menu translations
{
{
"zephyr_ortho_program": "QERMID - Orthopédie",
"zephyr_ortho_knee_project": "Orthopride genou",
"zephyr_ortho_knee_t1_dcd": "Primo-implantation"
},
{
"zephyr_ortho_program": "QERMID - Orthopédie",
"zephyr_ortho_knee_project": "Orthopride genou",
"zephyr_ortho_knee_t2_dcd": "Revision"
},
}
Get DCD field definition translations
Request example
GET {{baseURL}}/api/dcd/fields/translation?dcd-id={dcdId};dcd-version-id={dcdVersionId};language-id={languageId};field-id={fieldId}
Request parameters
- {dcdId} : Id of the menu dcd which we want to get all the translations.
- <optional parameter> {dcdVersionId} : If this parameter is not provided, lastest one is assumed.
- <optional parameter> {languageId} : language id for the menu options example results. If this parameter is not provided, default language will be English. Current permitted values:
- en: English
- nl: Dutch
- fr: French
- <optional parameter> {fieldName} : Id of field which we want to get the translation from
Request response
Example 1: languageId = "en" - English language fields translations
{
"author_group": "Authors group",
"CD_SLEEVE_LON": "Long sleeves",
"TX_TTL_UNT": "Unit",
"TX_CTNT_BEF_CONT_VEN_ARTER": "Before venous/arterial contact",
"CD_NAIL_DIRT": "Dirty nails",
"CD_PLSH": "Nail polish",
"TX_NHH": "No hand hygiene",
"CD_SITE": "Site number",
"CD_NAIL_EXT": "Nail extensions",
"TX_TTL_STDY": "Study design",
"CD_WATCH": "Wearing a watch",
"D_OBS": "Observation date",
"T_STP_OBS": "Time end observation period",
"CD_OPT_MDL": "Participation module optional",
"CD_SPLTY": "Specialty of the ward",
"CD_PROF": "Profession",
"TX_TTL_OBS": "Observations",
"CD_NAIL_LON": "Long nails",
"CD_RING": "Wearing a ring"
}
Example 2: languageId = "nl" - Dutch language fields translations
{
"author_group": "Auteursgroep",
"CD_SLEEVE_LON": "Lange mouwen",
"TX_TTL_UNT": "Eenheid",
"TX_CTNT_BEF_CONT_VEN_ARTER": "Voor contact met intravasculair stelsel",
"CD_NAIL_DIRT": "Vuile nagels",
"CD_PLSH": "Nagellak",
"TX_NHH": "Geen handhygiëne",
"CD_SITE": "Campus nummer",
"CD_NAIL_EXT": "Kunst-nagels",
"TX_TTL_STDY": "Study design",
"CD_WATCH": "Het dragen van polshorloge",
"D_OBS": "Observatiedatum",
"T_STP_OBS": "Tijdstip einde observatieperiode",
"CD_OPT_MDL": "Deelname optioneel module",
"CD_SPLTY": "Specialiteit van de dienst",
"CD_PROF": "Beroepsgroep",
"TX_TTL_OBS": "Observaties",
"CD_NAIL_LON": "Lange nagels",
"CD_RING": "Het dragen van ring"
}
Example 3: languageId = "fr" - French language fields translations
{
"author_group": "Groupe d’auteurs",
"CD_SLEEVE_LON": "Longues manches",
"TX_TTL_UNT": "Unité",
"TX_CTNT_BEF_CONT_VEN_ARTER": "Avant contact veineux/artériel",
"CD_NAIL_DIRT": "Ongles sales",
"CD_PLSH": "Ongles vernis",
"TX_NHH": "Pas d’hygiène des mains",
"CD_SITE": "Numéro du site",
"CD_NAIL_EXT": "Ongles artificiels",
"TX_TTL_STDY": "Study design",
"CD_WATCH": "Le port d’une montre",
"D_OBS": "Date de l’observation",
"T_STP_OBS": "Heure fin de la période d’observation",
"CD_OPT_MDL": "Participation module optionel",
"CD_SPLTY": "Spécialité du service",
"CD_PROF": "Profession",
"TX_TTL_OBS": "Observations",
"CD_NAIL_LON": "Ongles longs",
"CD_RING": "Le port d’une bague"
}
Search for SADMI medical devices
Request example
GET {{baseURL}}/api/externalreference/sadmi?
notificationCode={notificationCode};
name={name};
reference={reference};
distributorName={distributorName};
manufacturerName={manufacturerName};
classificationCode={classificationCode}
Request parameters
- <optional parameter> {notificationCode}
- <optional parameter> {name}
- <optional parameter> {reference}
- <optional parameter> {distributorName}
- <optional parameter> {manufacturerName}
- <optional parameter> {classificationCode}
Request response
{
{
"NotificationCode": "First Notification Code Example",
"Name": "First Name Example",
"Reference": "First Reference Example",
"URL": "http://first.url.example",
"Distributor": {
"NotificationNumber": "First Notification Number Example",
"Name": "First Distribuitor Name Example"
},
"Manufacturer": {
"Name": "First Manufacturer Name Example",
"CountryCode": {
"value": "First Country Code Value Example",
"standard": "First Country Code Standard Example"
},
"CountryName": {
"value": "First Country Name Value Example",
"lang": "First Country Name Lang Example"
}
},
"Classification": {
"Code": "First Classification Code Example",
"Description": "First Classification Description Example"
},
"State": {
"Name": "First State Name Example",
"ValidFrom": "First State ValidFrom Example",
"ValidTo": "First State ValidTo Example"
}
},
{
"NotificationCode": "Second Notification Code Example",
"Name": "Second Name Example",
"Reference": "Second Reference Example",
"URL": http://second.url.example,
. . . . .
},
. . . . .
}
Adding repeatable blocks to payload
Example
"TX_TTL_IMPLANT_DATA": [
{
"CD_IMPLANT": "127909",
"CD_PAC_SADMI_NOTIFIC": "000017811475",
"CD_IMPLANT_CAT": "",
"CD_IMPLANT_PRD_NM": "",
"TX_IMPLANT_PRODUC": "",
"TX_IMPLANT_DSTRBTR": "",
"TX_IMPLANT_DESC": ""
},
{
"CD_IMPLANT": "127910",
"CD_PAC_SADMI_NOTIFIC": "",
"CD_IMPLANT_CAT": "127906",
"CD_IMPLANT_PRD_NM": "productnaam",
"TX_IMPLANT_PRODUC": "fabrikant",
"TX_IMPLANT_DSTRBTR": "verdeler",
"TX_IMPLANT_DESC": "opmerking product"
}
],3. MDM Field description - DB Model
3. MDM Field description - DB Model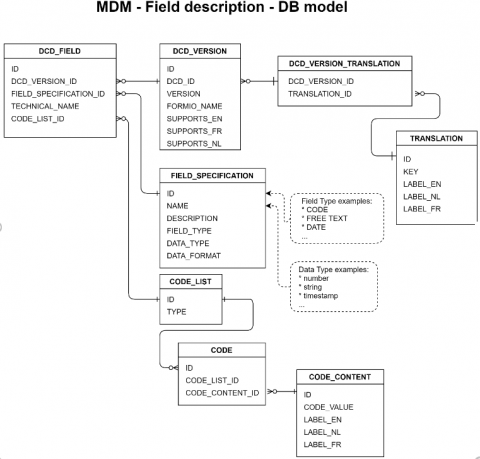
4. Swagger API
4. Swagger APIThe Swagger API is available on the local installation of HD4DP2.0. Go to https://<hd4dp2_url>/proxy/api-docs.html.
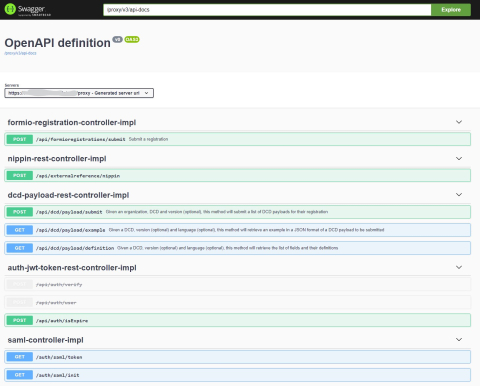
Some API calls are password protected (S2S api). If you want access to this API, you can request the user/password by creating a ticket in our servicedesk.
Attention: The baseURL to be used has to following built: https://<hd4dp_url>/proxy
5. Postman collection
5. Postman collectionPostman collection HD4DP2.0 can be downloaded here.
Attention: The baseURL to be used has to following built: https://<hd4dp_url>/proxy
HD4DP v2 CSV Upload
HD4DP v2 CSV UploadIntroduction
This page explains the functioning of the CSVUploader feature. The CSVUploader feature is aimed to do a bulk upload of records: by filling a csv file, one record per row represents one submission so a user can fill as much records as needed.
Training
Below you can review the HD4D¨2 CSV Upload training organized by healthdata.be:
Architecture
The CSVUploader is located under hd-connect/csvuploader. It uses both hd-connect-csvuploader and hd-connect-proxy modules.
The general architecture of the CSVUploader is explained in the sequence diagram below.
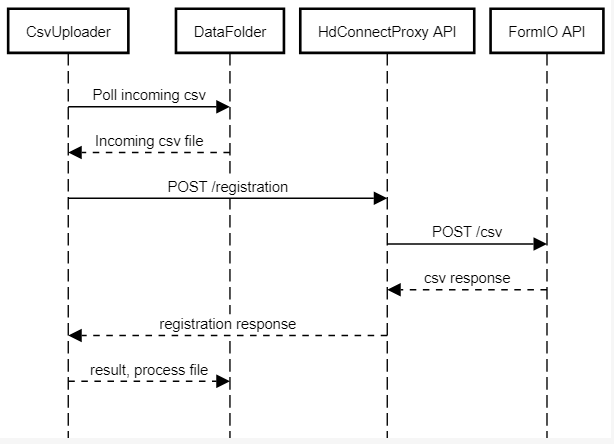
Third-party libraries and frameworks
- Apache Camel: https://camel.apache.org/
- Spring Boot
Testing and working
- The CSVUploader creates a folder at root level (SFTP for end-user, or hd-all for developer) that contains a subfolder per existing organisation.
- The CSVUploader will poll with a delay of 1 min, process the csv file and then create 3 folders:
- ARCHIVE folder: contains the source csv file.
- RESULTS folder: contains the results of processing the csv file. This file contains the specified data, and the final status of the processing: Success or Error. If an error occurred, the error message is displayed. For multiple uploads, the result is added to the end of the result file each time.
- ERROR folder: this folder is created if the csv test file was not processed, due to an I/O error (file corrupted, not found etc.). So for now, only technical errors are caught and the source csv file is moved to that folder instead of the ARCHIVE folder. In principle, this folder should contain any result that is an error. The RESULT folder should only contain results that end with a SUCCESS status.
Formats
Some formats are specific:
- Dates: should be dd/mm/yyyy
- Boolean: true / false
- Codes: the value of the code (not the translation)
- Multi codes: there is only one column per field. So when a select box is set as multiple, values have to be separated by a "|". e.g.: 68452|68453|68454
- Repeatable blocks: in some DCDs, a complete block of fields is repeatable. In that case, value have to be separated by a ";"
- e.g.: A block is containing 3 fields: A (Lob), B (Type klep) and C (Aantal kleppen)
Examples:
In case of a multiple choice:
Example of a multiple choice as presented in the DCD:

As presented in the form:
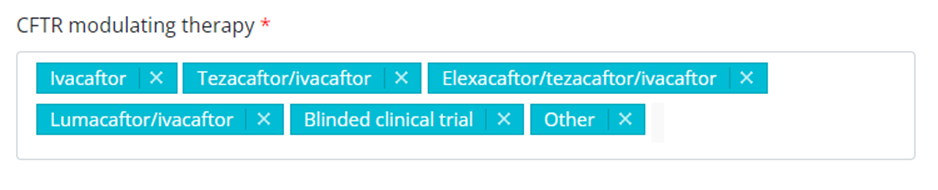
For reporting fields with a multiple choice formatting, the selected answers can be reported separated by the pipe symbol (|).
Example of a multiple field reporting:
cftr_modulating_therapy_1;
1|2|3|4|5|6;
In case of a repeatable block
Example of a repeatable block as presented in the DCD:

As presented in the form:
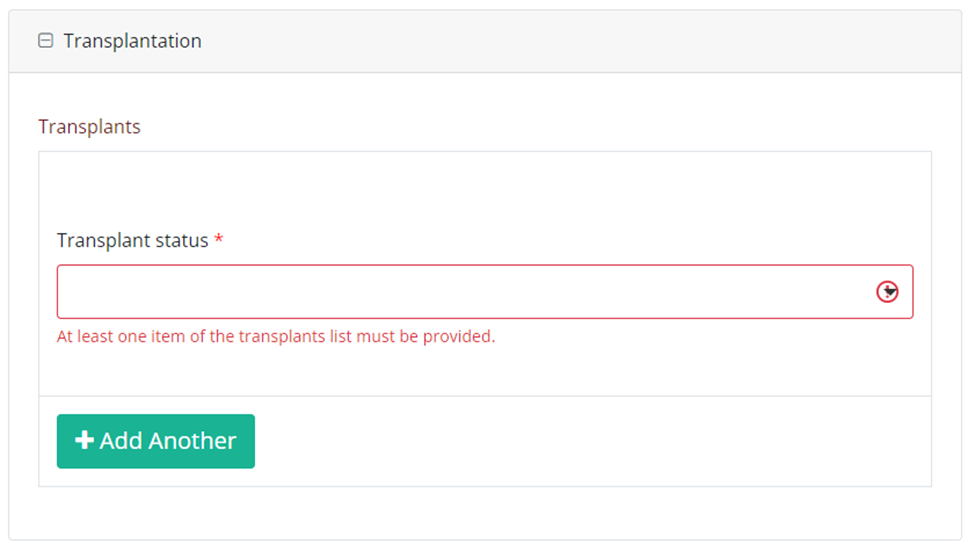
Manually enter the repeatable fields as shown in the following template: field_category|<index>|field.
Example of a repeatable block reporting:
Repeatable 1:
Transplants
transplant_status
Repeatable 2:
Transplants
transplant_status
becomes:
transplants|0|transplant_status;transplants|1|transplant_status
Process to upload a CSV file to submit DCD registrations
Process to upload a CSV file to submit DCD registrationsDocumentation for CSV Upload on Architecture 2.0
Description of the service
The CSV Upload functionality gives the possibility to import multiple parameters from a set of patients in one time into HD4DP 2.0. The csv file is based on an extract of the electronic patient files and/or other local databases.
Currently there is no user interface in HD4DP 2.0 for uploading csv files. If a data provider wants to upload a csv file, the file has to be dropped at a specific location. These files will be picked up and processed periodically. The files should be “final”, meaning that no application is writing to them. The pick-up location will be identical for all registries.
Pre-registry handling will be based on a naming-convention of the csv file.
HOW TO: Upload data using CSV Upload
Steps To Upload data
1. Prepare the csv file (example file in this section)
- Extract the csv file from the electronic patient files and/or other local databases.
- Author group, Author and Coauthor:
- When the Author Group, Author and Coauthor have been left out in the csv file, the default Author group, Author and Coauthor will be used automatically.
- When the desired Author Group, Author and Coauthor are specified in the csv file, the following headers TX_AUTHOR_GR, TX_AUTHOR and TX_COAUTHOR must be added to the csv file with their values respectively.
Example:
TX_AUTHOR_GR;TX_AUTHOR;TX_COAUTHOR
Test group;test@sciensano.be;test@sciensano.beNote:
The Author group, Author and Coauthor must exist and are well configured at the back-end of the system. TX_AUTHOR_GR can be a string that identifies the Author group to which this Author belongs. Commonly, the first name and last name are used to identify the TX_AUTHOR_GROUP. Be sure to avoid leading and trailing spaces when entering the Author group value.
- Similarly, add the field name 'STATUS' in capitals in an additional column. Add the value 'draft' in case a manual submission of the record is requested.
If not, the record will be submitted without manual intervention.
- Adding separators to a NISS number:
It is not necessary to add separators in a NISS number when uploading a file using CSV Upload. You can fill out the NISS number both with or without separators. E.g.: 85.04.02-169.32 or 85040216932.
Example:
- Make sure the name of the csv file has the correct format:
HD_DCD_submcsv_HDBPnumber_HDBPabbreviation_versionnumber_versionreleasedate
So FOR EXAMPLE for Orthopride Knee - Primo Implantation the format would be:
HD_DCD_submcsv_HDBP0288_Orthopride_Knee_Primo_02_20230505.csv
EXAMPLES:
- Please find below a HEADER file for Orthopride Knee - Primo-implantation:
- Please find below an example file for Orthopride Knee - Primo-implantation:
Disclaimer: The example files above are only provided as a guideline and do not contain real life data.
2. Uploading the CSV File
Step 1: Open the sftp tool like WinSCP

Step 2: Get the credentials (Host name, Port number, User name and Password) from the IT department of the Data Provider, to log on to the sftp server located on the Data Provider side.
Step 3: Fill in the credentials into the Login screen and click on Login to be able to access the different upload folders:
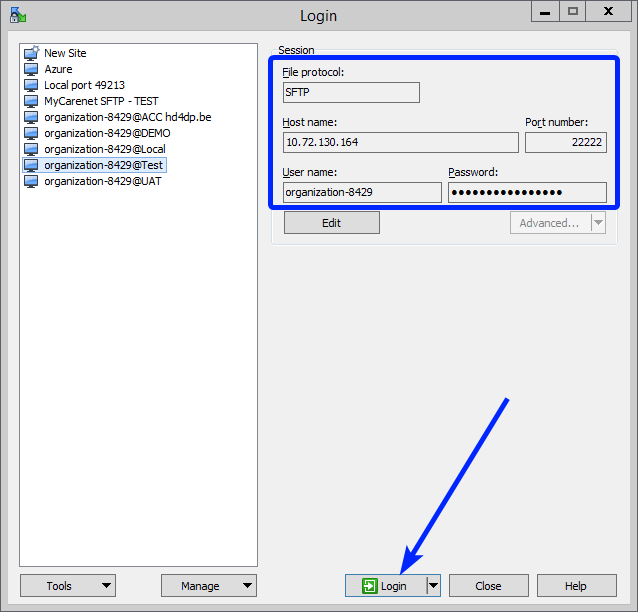
Note: a warning might be given, just click on Update
Now the CSVUpload folder structure is displayed on the right-hand side panel:
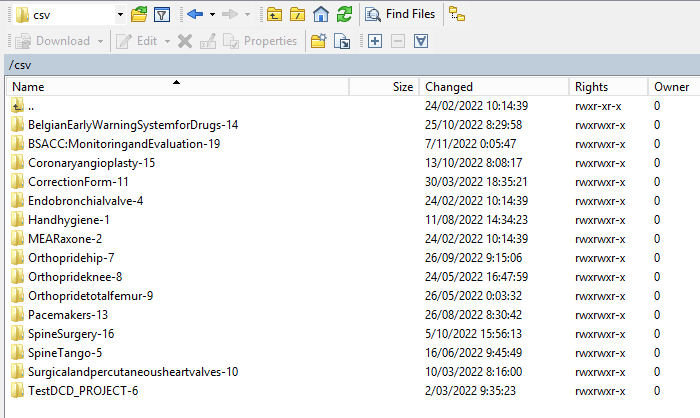
Step 4: Select the project folder Orthoprideknee-8 and open it by double-clicking on it:
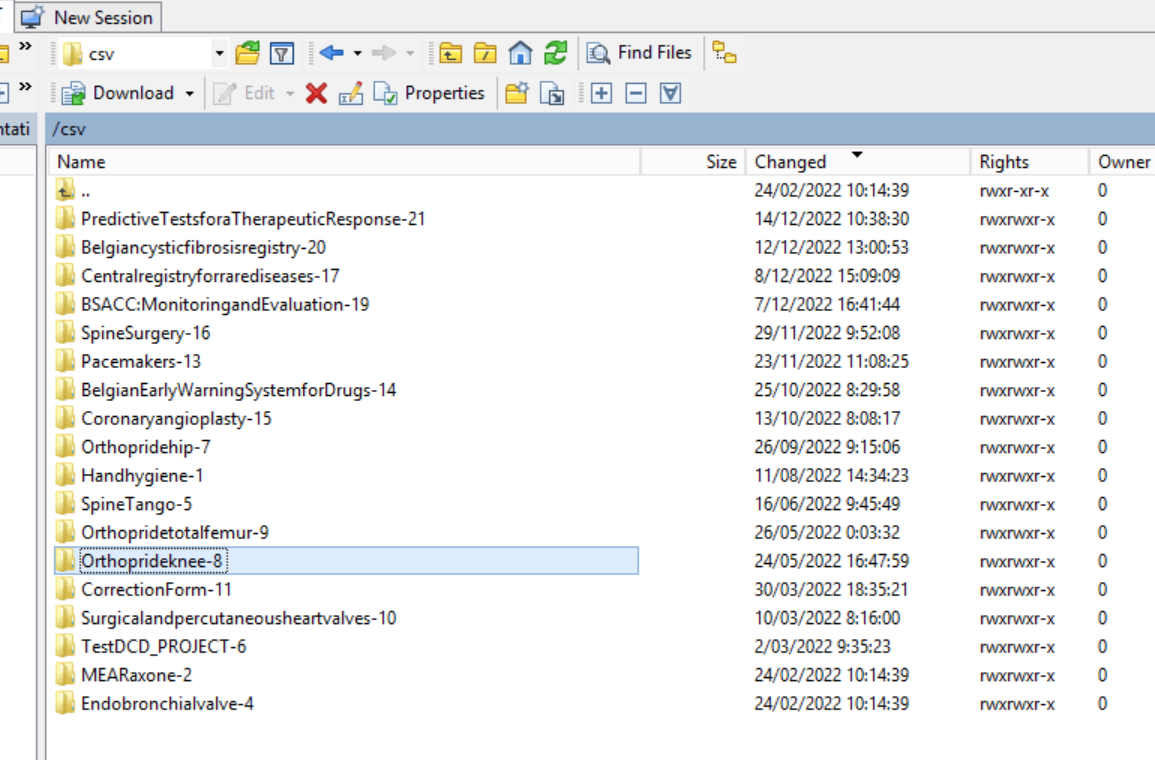
Step 5: Double-click on the DCD folder to open it:
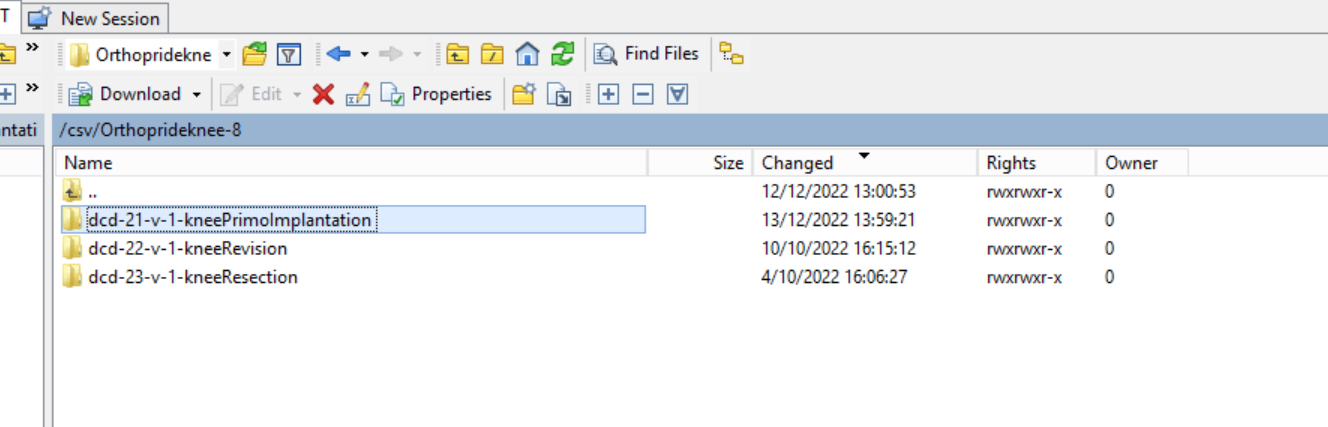
Step 6: Now go to the folder on the left-hand side panel where the CSV file to be uploaded is located:
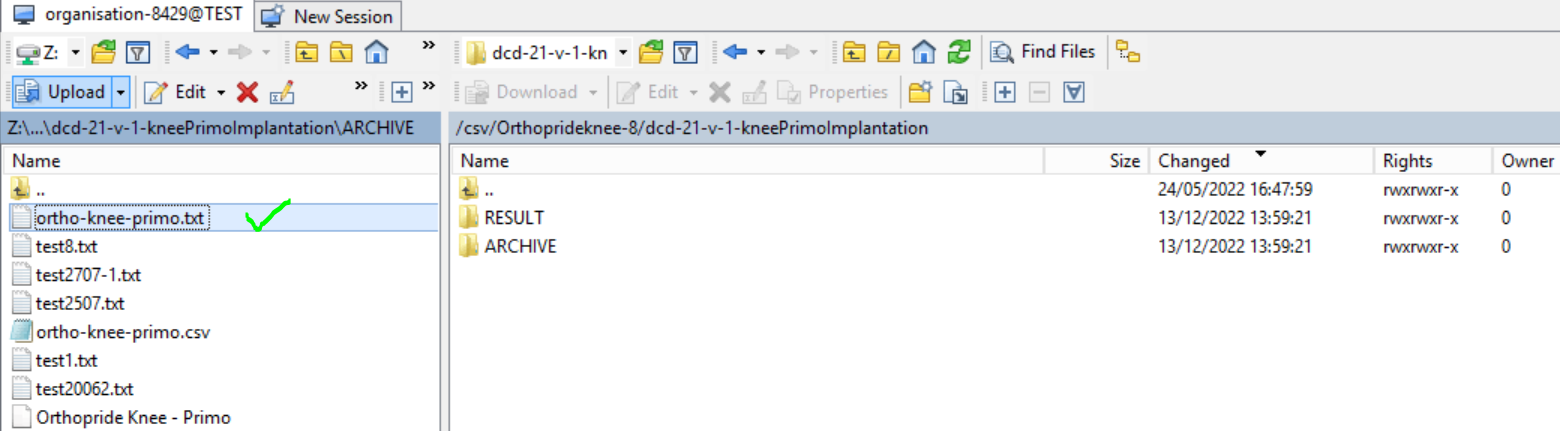
Step 7: Drag the CSV file to be uploaded from the left-hand side panel into the folder on the right-hand side panel:
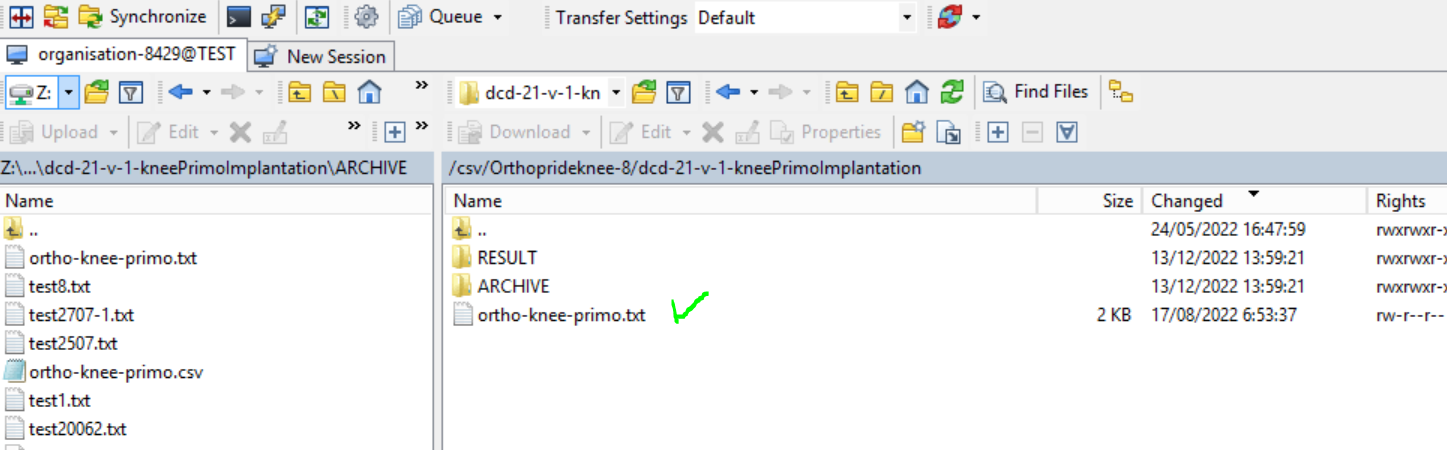
Step 8: Wait until the polling system of the CSV Uploader has picked up the CSV file and has processed it.
Once the CSV file has been processed it will disappear from the folder, when we refresh the page manually!
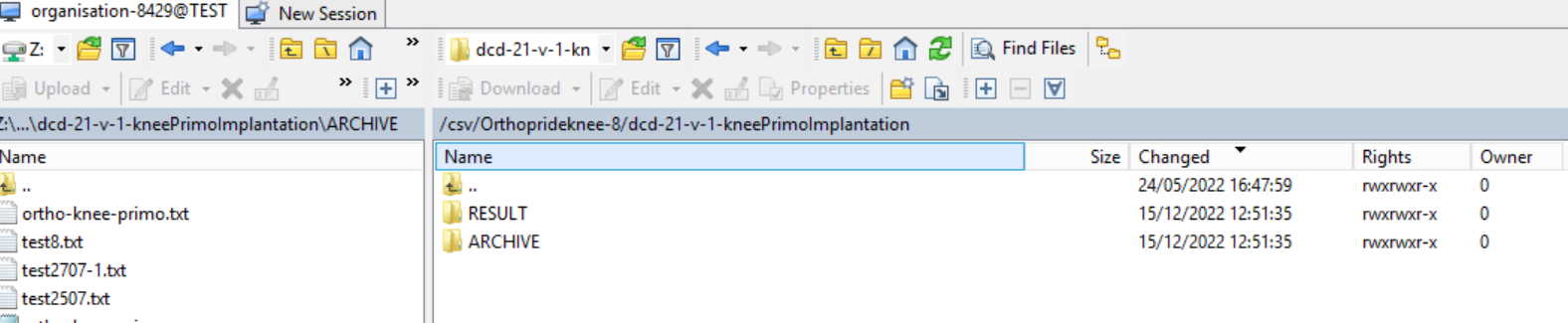
3. Validate CSV Upload
Once the CSV file has been processed 3 folders will be created (if they haven't been created already) in the DCD folder:
- ARCHIVE (after a csv file has been processed, the original csv file will be saved in this folder)
- RESULT (when the csv file is placed in this folder, it means that the csv file has been processed, a file will be created or (or appended, if the file already existed) with the result of the upload of the csv file).
All the errors that are described in th this file are business related, which means that they are technically correct, but in violation with the business rules or contains wrong values for that field. - ERROR (when the csv file is placed in this folder, it means that a technical error has occurred like the csv file contained erroneous formatting. The csv file won't get processed and an error file will be created with the errors and reason why the csv file couldn't be processed)
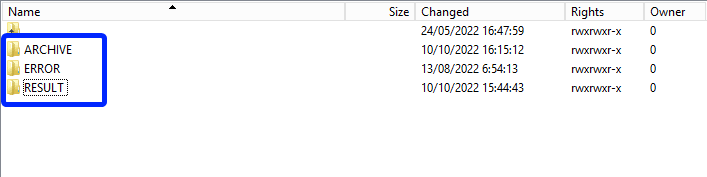
3.1 Validation of the CSV Upload via sftp tool:
Step 1: Double-click on the ERROR folder to open it, click on the refresh button and verify that there is no error file present.
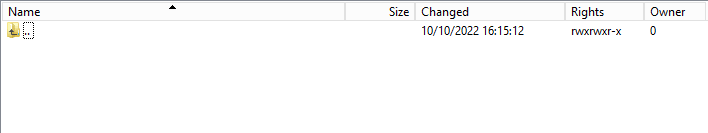
Step 2: Return to the DCD folder. Now double-click on the RESULT folder to open it, click on the refresh button and verify that the result file is present.
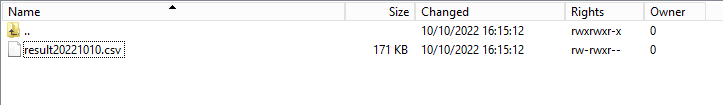
Step 3: Double-click on the result file to open it.
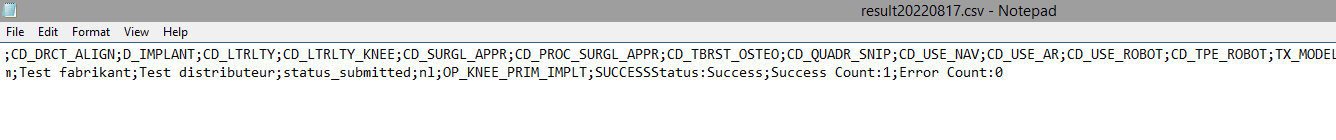
Step 4: If there are multiple records in the result file, scroll to the entry of the current csv upload by looking at the upload date (Started at dd/mm/yyyy hh:mm).
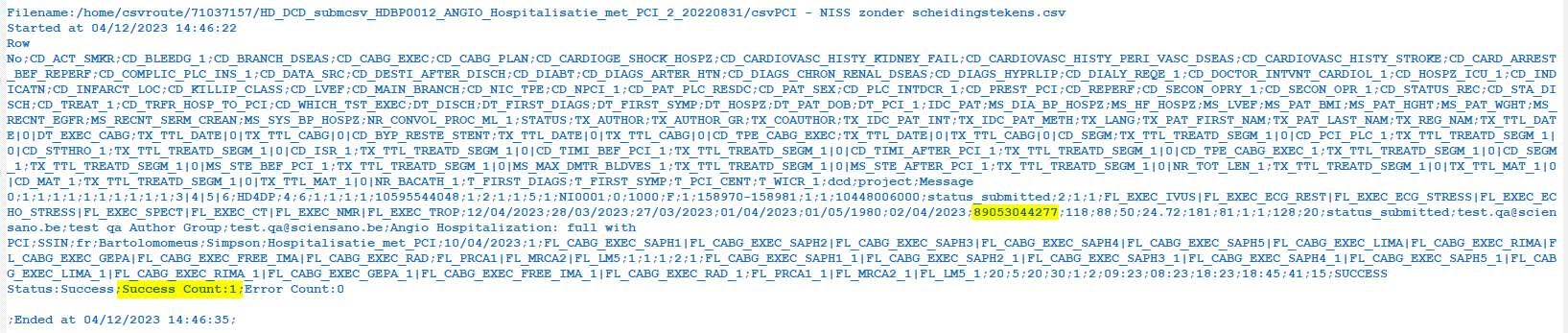
Verify the result file that the upload was successful by searching for the word SUCCESS and having a look at the Status. This Status must contain: Success;Success Count:1;Error Count:0
Step 5: If there are multiple records in the result file, scroll to the entry of the current csv upload by looking at the upload date (Started at dd/mm/yyyy hh:mm).
Verify the result file that the upload was successful by searching for the word SUCCESS and having a look at the Status. This Status must contain: Success;Success Count:1;Error Count:0
3.2 Validation of the CSV Upload via HD4DP 2.0:
Step 1: Open the web application HD4DP 2.0.
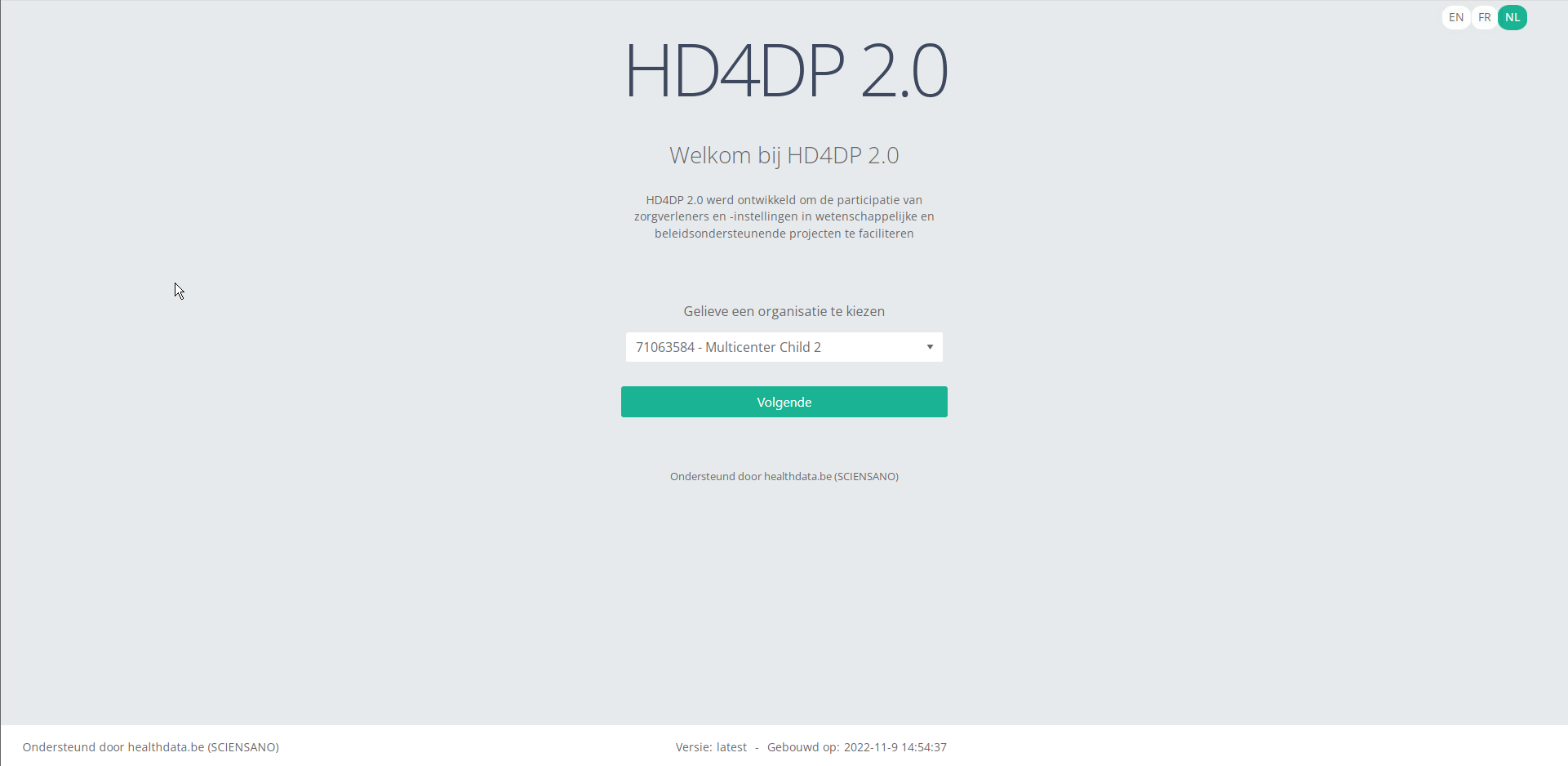
Step 2: Select the concerned organization in the dropdown list and click on Volgende (Next)
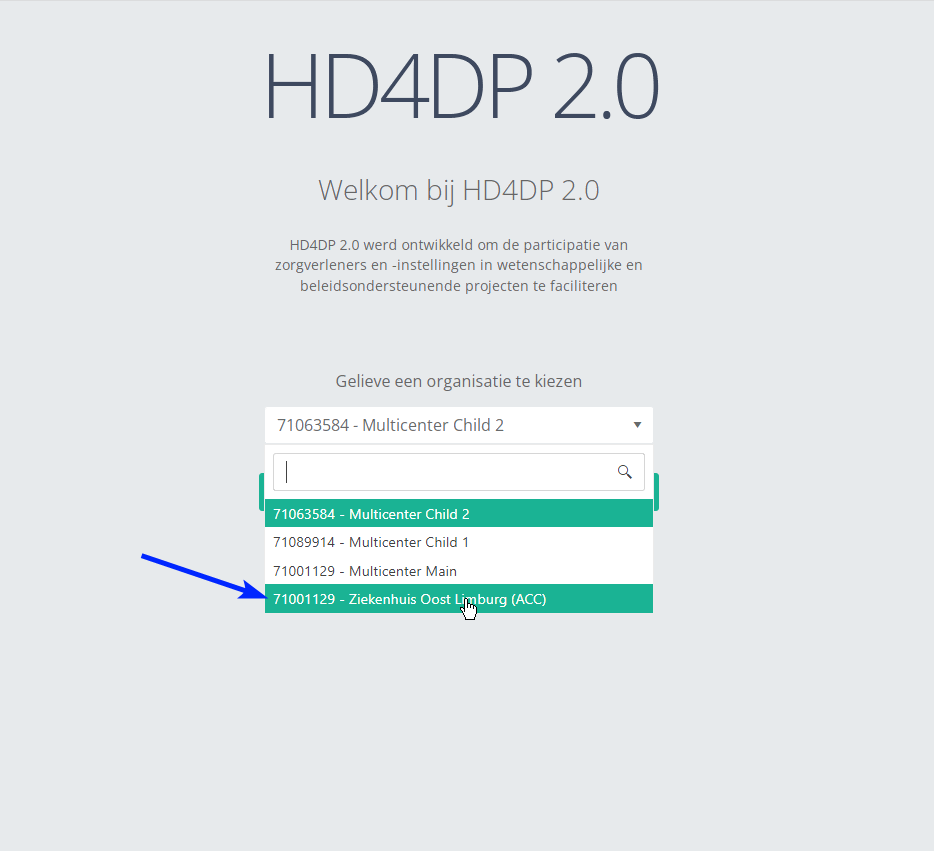
Step 3: Fill in the username and password, that has been provided by your IT Department or Healthdata team, and click on Log in to access the HD4DP 2.0 application.
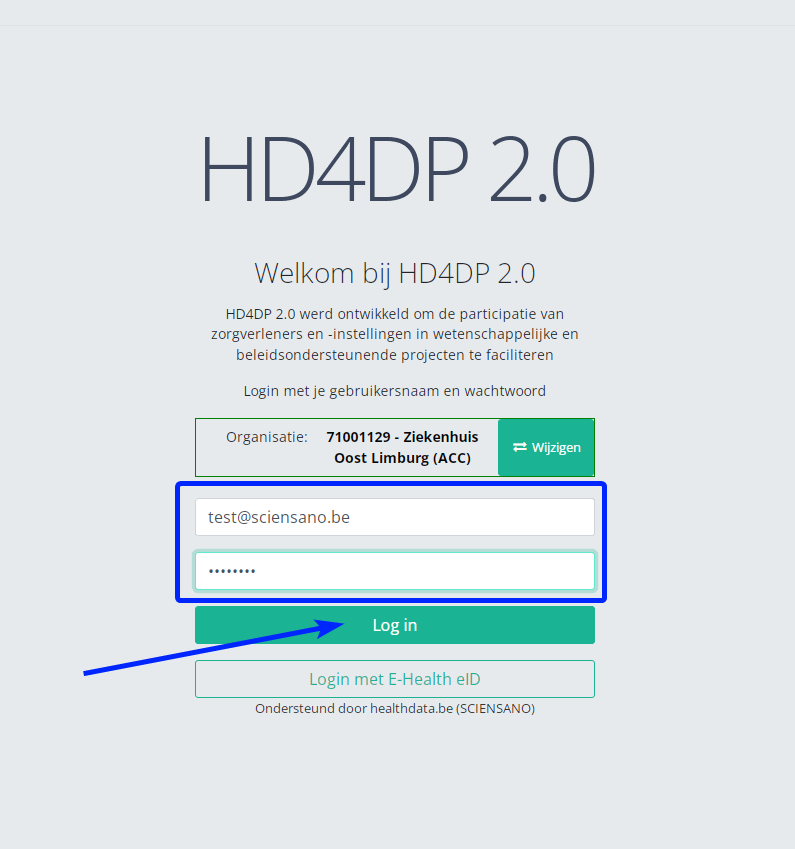
Step 4: Navigate in the menu on the left-hand side panel to the desired DCD:
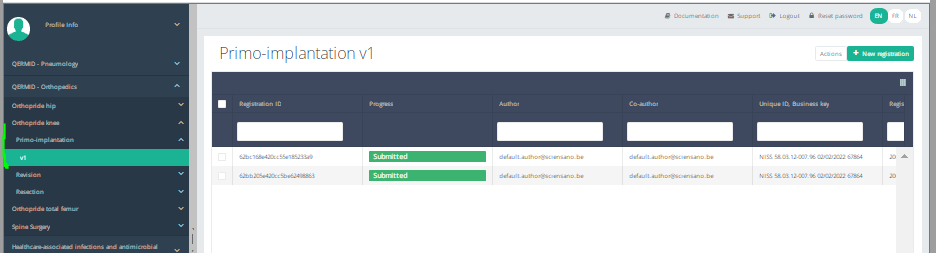
Step 5: Check that the uploaded DCD(s) is/are displayed in the DCD overview
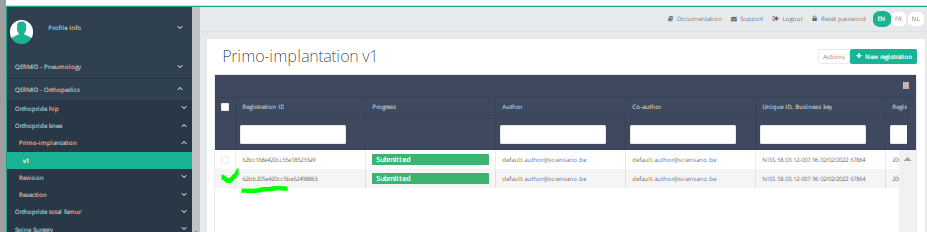
HD4DP v2 MyCareNet
HD4DP v2 MyCareNetHD4DP v.2 permits the administrative obligation of reporting to the insurance institutions. The limited necessary data are sent via HD4DP v.2 to the MyCarenet interface of the National Intermutual College (NIC). This transmission of nominative data occurs in parallel with the transmission of pseudonymized data to the healthdata.be platform.
Two options are available to enable the transmission of data from HD4DP v.2 to the National Intermutualist College (NIC):
This documentation is being updated regularly. We try to provide as correct, complete and clear as possible information on these pages. Nevertheless, if you see anything in the documentation that is not correct, does not match your experience or requires further clarification, please create a request (type : request for information) via our portal (https://sciensano.service-now.com/sp) or send us an e-mail via support.healthdata@sciensano.be to report this documentation issue. Please, do not forget to mention the URL or web address of the page with the documentation issue. We will then adjust the documentation as soon as possible. Thank you!Option 1: MyCareNet XML Export for [DCD name]
Option 1: MyCareNet XML Export for [DCD name]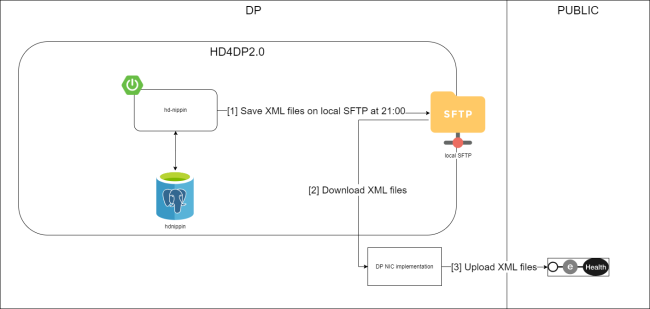
This configuration is by default active in HD4DP v2.
The files can be found on the SFTP share inside the directory /nippin/valid. The XML files can be downloaded with an SFTP client of your choice. The XML files you download can be sent with the MyCareNet component available in your organization.
Structure of the XML filename
The XML filename is a combination of different values: RIZIVnumber-random_uuid-inputReference.xml. The files are ready for use, thus contain the required fields. No additional editing is required.
How to link an XML file within the Nippin database
You can attach the XML files to the Web Service call to MyCareNet and sign the message with your eHealth P12 certificate. If you would like to search inside the Nippin database for the correct entry you can filter with the following query for the correct entries:
select count(*) from nippin_message where input_reference = '<inputReference>' and identification_value = '<RIZIVnumber>';
- Example: 11111130-6d59369f-11c8-4636-929a-86449f4dde3b-1701979200058.xml
- RIZIVnumber: 11111130
- random_uuid: 6d59369f-11c8-4636-929a-86449f4dde3b
- inputReference: 1701979200058
SFTP Connection
The server name and the SFTP credentials can be requested via our Service Portal
- Server: IP of HD4DP v.2 server
- Port: 22
- Username: (your SFTP credentials)
- Password: (your SFTP credentials)
- Path: /data/localsftp/upload/nippin (Upload is the home directory of the sftp user)
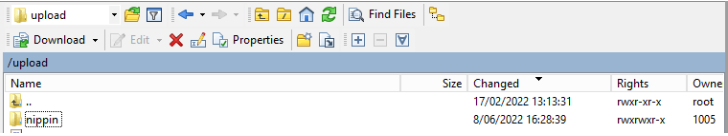
Option 2: MyCareNet integration in HD4DP v2
Option 2: MyCareNet integration in HD4DP v2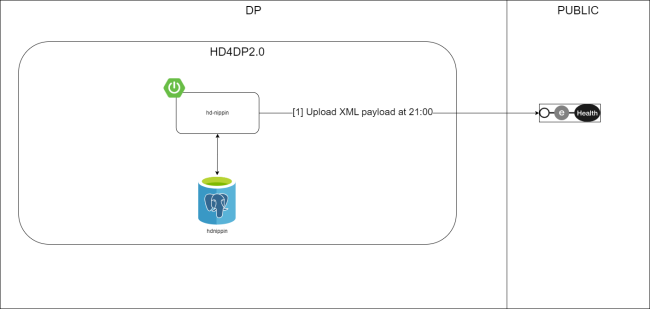
Four major actions are required in order to setup MyCareNet in HD4DP v.2:
- Whitelisting URLs
- Create certificate (ehealth_certificate.p12) with labels
- Upload eHealth certificate in HD4DP v.2
- Request credentials needed to upload the eHealth certificate
Whitelisting URL's
The following URLs must be whitelisted to communicate with MyCareNet and E-health. Without a direct connection from HD4DP v.2 server to these URLs, a registration to MyCareNet will not work.
https://prod.mycarenet.be:9443/*
https://services.ehealth.fgov.be/*
Create certificate (ehealth_certificate.p12) with labels
This part requires your organization's eHealth certificate. Create a certificate that has the name ehealth_certificate.p12 using the open source GUI KeyStore Explorer. Download and install the tool from https://keystore-explorer.org/
- Open your organization's eHealth certificate
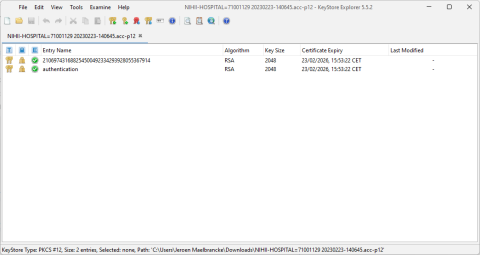
2. Export both certificates as a Key Pair
3. Open KeyStore Explorer, and create a new KeyStore
4. Select the type of the new KeyStore: PKCS#12
5. In the menu bar go to Tools -> Import Key Pair
6. Select the type of key pair import required:
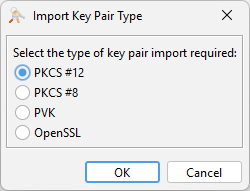
7. Browse the authentication certificate and fill in the Decryption Password
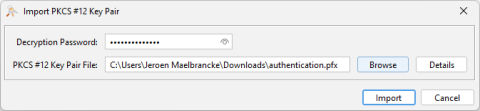
8. Import the authentication PKCS #12 Key Pair and give it the NIHII number as alias, ex 71001129
9. Click OK and give it a password that needs to be same for all imported certs in the P12 KeyStore
10. The Key Pair is imported Successfully
11. Repeat steps 5 to 10 for importing the serial number PKCS #12 Key Pair and give it the same serial number as alias
12. Repeat step 1 to 11 to add more NIHII-HOSPITAL P12 certificates to this KeyStore (Do not execute step 3 and 4 if you already created a new Keystore)
13. In the menu bar goto File-> Save All
14. Set the KeyStore Password and give it a password that needs to be same for all certificates

15. Click OK and save the KeyStore asehealth_certificate.p12
16. Click save and the P12 for HD4DP is created
Upload eHealth certificate in HD4DP v.2
The filename of your P12 certificate must be ehealth_certificate.p12
Server: IP of HD4DP v. 2 server
Username: (your SFTP credentials)
Password: (your SFTP credentials)
Path: /data/localsftp/upload (home directory of thesftpuser)
File: ehealth_certificate.p12
Request credentials needed to upload the eHealth certificate
The server name and the sftp credentials can be requested via our Service Portal. The password of your P12 certificate can be delivered to healthdata.be either via a secure password sharing tool of your choice or via Belnet Filesender to hd-architecture-2@sciensano.be. You can request a Belnet Filesender voucher via our Service Portal as well.
Architecture 2.5
Architecture 2.5 Bart.Servaes Sat, 01/06/2024 - 15:51Uniformed naming convention
Uniformed naming conventionChange name:
Create DCD formio naming structure using a general overall naming convention of healthdata projects.
Change Description:
The change is to refactor the formio DCD naming to have a structure as:
HDBPnumberHDBPabbreviationdcdname/abbreviationversionnumber
Where the abbreviation will be used as seen in the column “TX_PROJ_BUS_ABBREV” of the document below, that will be a reference to fill the MDM:
Example 1:
So for the formio DCDs of the project “Mandatory registration of spine surgery” (full name) it would be something like:
HDBP0240_SPINE_1 and because there is only 1 dcd for the project we do not use a dcd name.
Example 2:
So for the formio DCDs of the project “Pneumology Endobronchial valve” (full name) it would be something like:
HDBP0231_ZEPHYR_ZEPHYR_REPLAC_1
HDBP0231_ZEPHYR_ZEPHYR_PRIM_IMPLT_1
Full example:
dcdname/abbreviation: will be the TX_REG_NAME of the DCD.
| form title | form name | form path | collection name |
| HDBP0240_SPINE | hdbp0240Spine1 | hdbp0240spine1 | HDBP0240SPINE1 |
| HDBP0231_ZEPHYR_ZEPHYR_PRIM_IMPLT | hdbp0231ZephyrZephyrPrimImplt1 | hdbp0231zephyrzephyrprimimplt1 | HDBP0231ZEPHYRZEPHYRPRIMIMPLT1 |
Sending code values instead of code IDs in S2S requests
Sending code values instead of code IDs in S2S requestsIn the following example we will demonstrate sending code values instead of code IDs. We want, for instance, to send a patient's country and zip code information, more specifically for someone who lives in Brussels, Belgium:
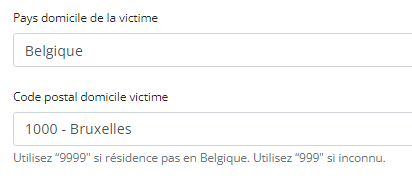
Currently, in the case of fields containing code list elements with S2S submissions, we send the code ID in the body of the message, like so:
"CD_CNTRY_RES": "130349",
"CD_POSTCODE": "4525",In CSV upload, however, we send the code value of the code object.
From the new Architecture 2.5 onward, we will do so too for S2S submissions using the API. So, for the example above, the new way of sending the same values will be as follows:
"CD_CNTRY_RES": "BE", "CD_POSTCODE": "1000",The image below shows the relationship between these fields in the database, in the case of the country:

And for the postal code:

The purpose of this change is to stop sending the code IDs in System-to-System and start sending the code values, just like we are doing in the CSV upload. The performance improvements that go along with this result from the fact that the service that returns the code value for a given ID is done through a single request per submission to the MDM, thus saving a lot of time in calls to get these values.
Since the code value is not unique, we need to get the codelist ID of an element, given its key, from a given DCD version ID. This could be summarized for this example as follows:
The key CD_CNTRY_RES on DCD version ID 55 (BSACC Police And Public Prosecutor) has the codelist id 347.
The key CD_POSTCODE on DCD version ID 55 (BSACC Police And Public Prosecutor) has the codelist id 12.Code Lists and Code Values are agreed upon by IAT and the Researchers. These are sent to DC by IAT via the DCD specifications. A Code ID is the identification of a Code Value in MDM. So how is the translation done?
In the case of sending code IDs, the user calls our services to retrieve the code IDs through requests to our API. In these requests, the user always receives the complete code object, containing not only the code ID, but also the code value. Let’s have a look at an example for the country code (code list 347):
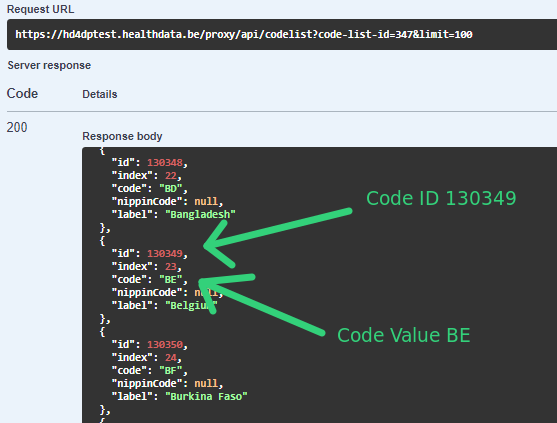
The image shows how we were sending the code ID 130349 in System-to-System and how we can send the code value “BE” in Architecture 2.5, the same way that was done already using the CSV Uploader.
In terms of code, if we were already calling the services to get the code ID, the only thing that is necessary is changing the returned value from CodeID to CodeValue from the same Code object that is returned.
FROM:
mappedCode = codeFieldService.getFieldCode(field.getFieldId(), codeContent).codeId();TO:
mappedCode = codeFieldService.getFieldCode(field.getFieldId(), codeContent).codeValue();That’s the only change needed in the mapping. All the rest remains exactly the same.
For more information, please check our documentation at 1. End-to-End process to submit DCD registrations | docs.healthdata.be
Architecture 2.0 and Architecture 2.5 proxies
Below you find an example of how the service to get a project-result is called in both proxies:
Architecture 2.0
https://hd4dp.acceptance.healthdata.be/proxy/api/installation/project-result?organization-id=120
[{"projectId":4,"dcdId":9,"dcdVersionId":9,"version":1,"formioName":"hdbp0231ZephyrZephyrPrimImplt1"}, {"projectId":4,"dcdId":10,"dcdVersionId":10,"version":1,"formioName":"hdbp0231ZephyrZephyrReplac1"}, {"projectId":4,"dcdId":11,"dcdVersionId":11,"version":1,"formioName":"hdbp0231ZephyrZephyrFllwup1"}, {"projectId":5,"dcdId":12,"dcdVersionId":12,"version":3,"formioName":"hdbp0062BelPstSpineTangoIntake1"}, {"projectId":5,"dcdId":13,"dcdVersionId":13,"version":3,"formioName":"hdbp0062BelPstSpineTangoConservativeTherapy1"}, {"projectId":5,"dcdId":14,"dcdVersionId":14,"version":3,"formioName":"hdbp0062BelPstSpineTangoPatientQuestionnaire1"}, {"projectId":5,"dcdId":15,"dcdVersionId":15,"version":3,"formioName":"hdbp0062BelPstSpineTangoSurgery1"}, {"projectId":6,"dcdId":16,"dcdVersionId":16,"version":1,"formioName":"hdbp0000TestTestDcd011"}, {"projectId":6,"dcdId":17,"dcdVersionId":17,"version":1,"formioName":"hdbp0000TestTestDcd021"}, {"projectId":7,"dcdId":18,"dcdVersionId":18,"version":1,"formioName":"hdbp0386OrthoprideHipOpHipPrimImplt1"}, {"projectId":7,"dcdId":19,"dcdVersionId":19,"version":1,"formioName":"hdbp0386OrthoprideHipOpHipRevis1"}, {"projectId":7,"dcdId":20,"dcdVersionId":20,"version":1,"formioName":"hdbp0386OrthoprideHipOpHipResec1"}, {"projectId":8,"dcdId":21,"dcdVersionId":21,"version":1,"formioName":"hdbp0288OrthoprideKneeOpKneePrimImplt1"}, {"projectId":8,"dcdId":22,"dcdVersionId":22,"version":1,"formioName":"hdbp0288OrthoprideKneeOpKneeRevis1"}, {"projectId":8,"dcdId":23,"dcdVersionId":23,"version":1,"formioName":"hdbp0288OrthoprideKneeOpKneeResec1"}, {"projectId":9,"dcdId":24,"dcdVersionId":24,"version":1,"formioName":"hdbp0048OrthoprideMegaOpMpPrimImplt1"}, {"projectId":9,"dcdId":25,"dcdVersionId":25,"version":1,"formioName":"hdbp0048OrthoprideMegaOpMpRevis1"}, {"projectId":9,"dcdId":26,"dcdVersionId":26,"version":1,"formioName":"hdbp0048OrthoprideMegaOpMpResec1"}, {"projectId":1,"dcdId":1,"dcdVersionId":30,"version":3,"formioName":"hdbp0025HhNsihHhPre3"}, {"projectId":1,"dcdId":2,"dcdVersionId":31,"version":3,"formioName":"hdbp0025HhNsihHhIo3"}, {"projectId":1,"dcdId":3,"dcdVersionId":32,"version":3,"formioName":"hdbp0025HhNsihHhPost3"}, {"projectId":10,"dcdId":27,"dcdVersionId":33,"version":1,"formioName":"hdbp0245TaviTaviImplt1"}, {"projectId":10,"dcdId":28,"dcdVersionId":34,"version":1,"formioName":"hdbp0245TaviTaviFllwup1"}, {"projectId":11,"dcdId":29,"dcdVersionId":35,"version":1,"formioName":"hdbp0000CorrectionForm1"}, {"projectId":13,"dcdId":34,"dcdVersionId":40,"version":2,"formioName":"hdbp0016PacemakerQermidPacemakerPrimoImplantation2"}, {"projectId":13,"dcdId":35,"dcdVersionId":41,"version":2,"formioName":"hdbp0016PacemakerQermidPacemakerAjoutRemplacementElectrode2"}, {"projectId":13,"dcdId":36,"dcdVersionId":42,"version":2,"formioName":"hdbp0016PacemakerQermidPacemakerRemplacement2"}, {"projectId":13,"dcdId":37,"dcdVersionId":43,"version":2,"formioName":"hdbp0016PacemakerQermidPacemakerExplantation2"}, {"projectId":13,"dcdId":38,"dcdVersionId":44,"version":2,"formioName":"hdbp0016PacemakerQermidPacemakerSuivi2"}, {"projectId":14,"dcdId":39,"dcdVersionId":45,"version":3,"formioName":"hdbp0019Bewsd3"}, {"projectId":15,"dcdId":40,"dcdVersionId":46,"version":2,"formioName":"hdbp0012AngioHospitalisatie2"}, {"projectId":15,"dcdId":41,"dcdVersionId":47,"version":2,"formioName":"hdbp0012AngioHospitalisatieMetPci2"}, {"projectId":15,"dcdId":42,"dcdVersionId":48,"version":2,"formioName":"hdbp0012AngioHospitalisatieMetFfr2"}, {"projectId":15,"dcdId":43,"dcdVersionId":49,"version":2,"formioName":"hdbp0012AngioHospitalisatieMetFfrEnPci2"}, {"projectId":15,"dcdId":44,"dcdVersionId":50,"version":2,"formioName":"hdbp0012AngioFollowUpNaPci2"}, {"projectId":16,"dcdId":45,"dcdVersionId":51,"version":1,"formioName":"hdbp0240Spine1"}, {"projectId":17,"dcdId":46,"dcdVersionId":52,"version":4,"formioName":"hdbp0008Crrd4"}, {"projectId":19,"dcdId":48,"dcdVersionId":54,"version":1,"formioName":"hdbp0242BsaccMeBsaccReprt1"}, {"projectId":19,"dcdId":49,"dcdVersionId":55,"version":1,"formioName":"hdbp0242BsaccMeBsaccFuPolce1"}, {"projectId":19,"dcdId":50,"dcdVersionId":56,"version":1,"formioName":"hdbp0242BsaccMeBsaccFuCont1"}, {"projectId":19,"dcdId":51,"dcdVersionId":57,"version":1,"formioName":"hdbp0242BsaccMeBsaccFuReferral1"}, {"projectId":19,"dcdId":52,"dcdVersionId":58,"version":1,"formioName":"hdbp0242BsaccMeBsaccFuMed1"}, {"projectId":19,"dcdId":53,"dcdVersionId":59,"version":1,"formioName":"hdbp0242BsaccMeBsaccFuPsychc1"}, {"projectId":20,"dcdId":54,"dcdVersionId":60,"version":9,"formioName":"hdbp0001Bcfr9"}, {"projectId":21,"dcdId":55,"dcdVersionId":61,"version":3,"formioName":"hdbp0078Pitter3"}, {"projectId":22,"dcdId":56,"dcdVersionId":62,"version":3,"formioName":"hdbp0051Becpr3"}, {"projectId":23,"dcdId":57,"dcdVersionId":63,"version":1,"formioName":"hdbp0037Epilabo1"}, {"projectId":24,"dcdId":58,"dcdVersionId":64,"version":1,"formioName":"hdbp0244RProfildRprofildTreat1"}, {"projectId":24,"dcdId":59,"dcdVersionId":65,"version":1,"formioName":"hdbp0244RProfildRprofildRenewal1"}, {"projectId":25,"dcdId":60,"dcdVersionId":66,"version":1,"formioName":"hdbp0274HartDefibDefibPrimImplt1"}, {"projectId":26,"dcdId":61,"dcdVersionId":67,"version":1,"formioName":"hdbp0056MvoMvoImp1"}, {"projectId":26,"dcdId":62,"dcdVersionId":68,"version":1,"formioName":"hdbp0056MvoMvoFu1"}, {"projectId":25,"dcdId":63,"dcdVersionId":69,"version":1,"formioName":"hdbp0274HartDefibDefibExpl1"}, {"projectId":25,"dcdId":64,"dcdVersionId":70,"version":1,"formioName":"hdbp0274HartDefibDefibRepl1"}, {"projectId":25,"dcdId":65,"dcdVersionId":71,"version":1,"formioName":"hdbp0274HartDefibDefibElect1"}, {"projectId":27,"dcdId":67,"dcdVersionId":72,"version":1,"formioName":"hdbp0000DvrForm1"}]Architecture 2.5
[{"projectId":19,"dcdId":49,"dcdVersionId":55,"version":1,"formioName":"hdbp0242BsaccMeBsaccFuPolce1"}]As we can see, using proxy (Architecture 2.0) we have all projects from organization 120, while proxy (Architecture 2.5) returns only one element in its response. This is because the DCD filter in proxy (Architecture 2.5) is done in the database more efficiently, while in the proxy Architecture 2.0 the same filter was done in the code after the service call.
So, the filter is not used in the first call and if Architecture 2.0 calls the same service in Architecture 2.5 without the filter (https://hd4dp.acceptance.healthdata.be/proxy/api/installation/project-result?organization-id=120) it will get exactly the same result.
Architecture 2.0 and Architecture 2.5 proxies tests
CSV Uploader
For the CSV Upload, nothing has changed. So the same inputs are valid for both architectures. Here are the example of some successful tests:
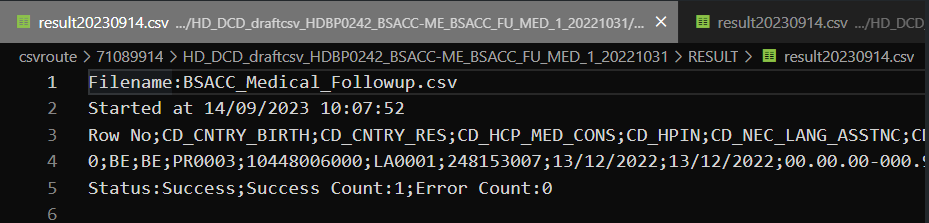
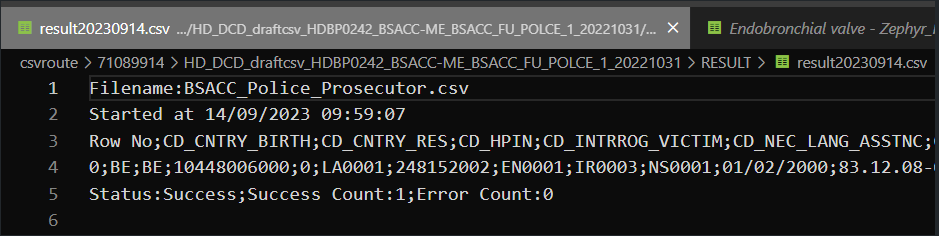
System-To-System
For S2S, as already mentioned, the only difference is that Architecture 2.0 sends Code IDs while Architecture 2.5 sends Code Values. Here are the same tests in both architectures:
Proxy (Architecture 2.0):
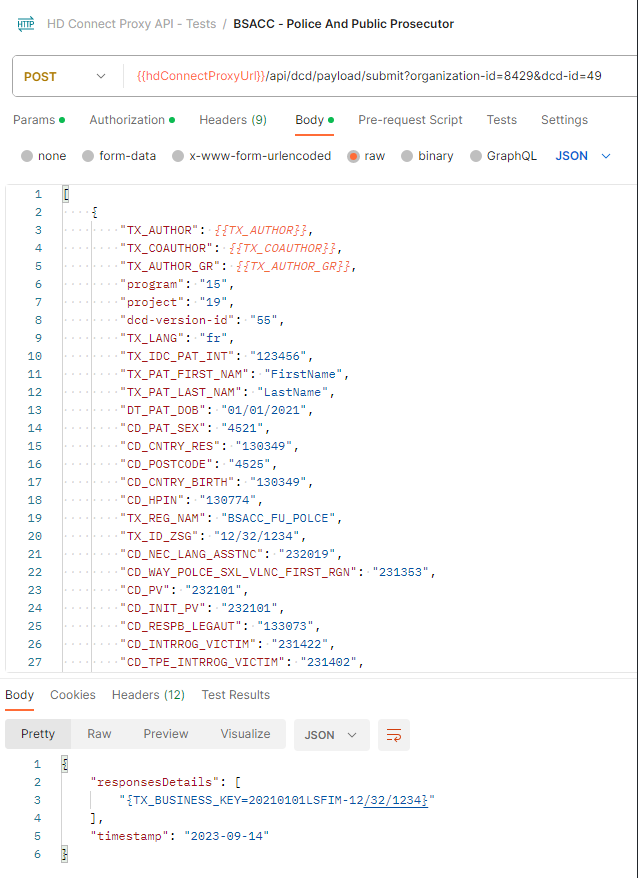
Proxy (Architecture 2.5):
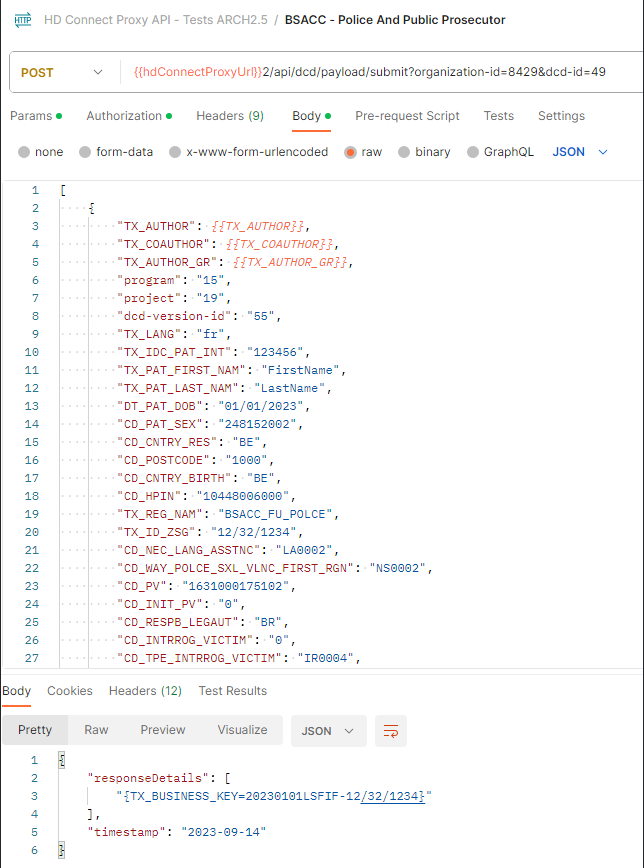
As we can see, we have the same request, where the first sends Code IDs and the second sends Code Values; both return a successful response.
Front-end
In the front-end (GUI) everything is received normally as the same request was sent twice. After all, S2S will always send the same request with Code IDs to FormIO.
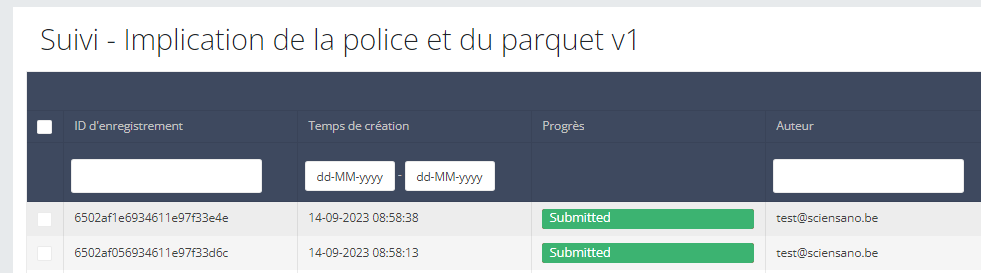
Data Warehouse
Once again, all the information arrives seamlessly, due to the fact that the FormIO validation already happened in the previous step:

First call (Architecture 2.0):
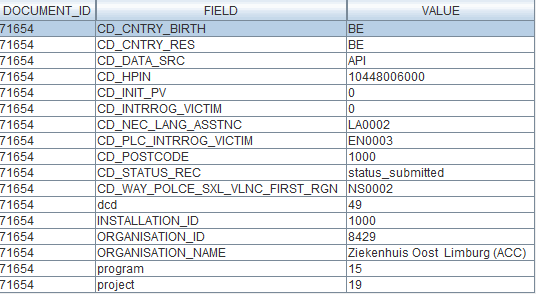
Second call (Architecture 2.5):
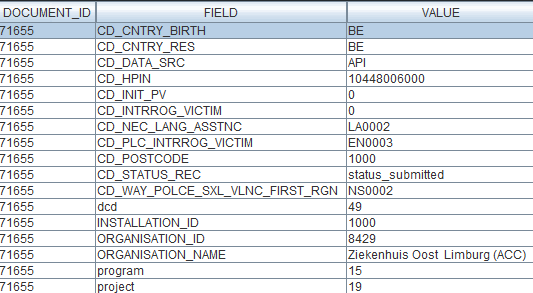
MDM Mapping of billing codes for MyCareNet
MDM Mapping of billing codes for MyCareNetNote: The below described mapping solution only applies to the study project of PACEMAKER.
In Architecture 2.0 the DCD fields related to billing codes for MyCareNet do not contain the billing code as their code value, instead this is just a numerical value, and the code value is only displayed in the label, e.g.:

When then sending key values to localdwh and dwh, the submission will contain a key value as follows:

When generating a message for MyCareNet, however, this same numerical value is used in the message payload. Keeping these numerical values, will generate MyCareNet messages with a incorrect billingcode, in this case <BillingCode>2</BillingCode>.

In Architecture 2.5 an MDM Mapping of billing codes for MyCarenet has been implemented. The value for the key will no longer be a numerical value such as 1,2,3… but an actual billing code. To facilitate this requirement, a new column has been introduced in the MDM to associate the billing code with the correct 13 character value, such as "182932-182943". This billing code will be accepted by MyCareNet.
Databases
Databases Bart.Servaes Wed, 02/14/2024 - 17:32Retrieve data from the local database of HD4DP v2
Retrieve data from the local database of HD4DP v2Warning
The person with the login for the local database of "HD4DP v2 local" has access to all the data stored in the database. This means that the personal data of the patients will be VISIBLE to that user.Requirements
URL Local DWH Database: postgresql://<server_ip>:5432/localdwh. If this is not the case, the IT department hosting HD4DP v2 needs to open the port and allow traffic to this port.
URL NIPPIN Database: postgresql://<server_ip>:5432/nippin
Username/Password: The service desk of healthdata.be will forward, via a secure link, the username and password.
Client: Download one of the clients that support PostgreSQL . A list is available here.
IP network/subnet: Provide us with the IP network/subnet from where you will contact the database. The database only accepts incoming traffic of known IP networks/subnets. example: 0.0.0.0/32
Granted privileges
| database | user | privileges |
| localdwh | dpuser | CONNECT/local_dwhmessage:SELECT/local_dwhmessage_key_value:SELECT/local_dwhmessage_key_value_plus:SELECT |
| nippin | dpuser | CONNECT/nippin_message:SELECT/nippin_cleanup:SELECT |
"data_collection_name" in local database
Query examples
With the "data_collection_name" and the following information, you will be able to link multiple tables with each other.
- local_dwhmessage_key_value: Key value table with more information about the registration
- msg_document_id: document id of your message located in local_dwhmessage table
- document_id: document id of your registration
- local_dwhmessage: table where you can find all the registrations
- local_dwhmessage_key_value_plus: Extra table to define attribute type and value of a key value
- key_value_id: Key value id linked to the id of the local_dwh_message_key_value
- local_dwhmessage_key_value:
"local_dwhmessage_key_value" column "msg_document_id" refer to the "document_id" of "local_dwhmessage".
"local_dwhmessage_key_value_plus" column "key_value_id" refer to the id of "local_dwhmessage_key_value".
Query 1: Get all registrations from the last 15 days.
SELECT * from local_dwhmessage WHERE data_collection_name = 'add data_collection_name' and created_on > current_date - interval '15' day;Query 2: Get all registrations and key value.
SELECT * from local_dwhmessage as ldm left join local_dwhmessage_key_value as ldmkv on ldmkv.msg_document_id = ldm.document_id WHERE ldm.data_collection_name = 'add data_collection_name';Query 3: Get all registrations, key value and key value plus from.
SELECT * from local_dwhmessage as ldm left join local_dwhmessage_key_value as ldmkv on ldmkv.msg_document_id = ldm.document_id left join local_dwhmessage_key_value_plus as ldmkvp on ldmkvp.key_value_id = ldmkv.id WHERE ldm.data_collection_name = 'add data_collection_name';Query 4: Get all MyCareNet registrations, key value and key value plus.
SELECT value from local_dwhmessage as ldm left join local_dwhmessage_key_value as ldmkv on ldmkv.msg_document_id = ldm.document_id WHERE ldm.data_collection_name = 'add data_collection_name'and key = 'TX_REGN_CD';
select * from local_dwhmessage_key_value where msg_document_id in ( select msg_document_id from local_dwhmessage_key_value where key = 'TX_REGN_CD' and value = 'use value from first query');
select * from local_dwhmessage where document_id in ( select msg_document_id from local_dwhmessage_key_value where key = 'TX_REGN_CD' and value = 'use value from first query');
| Column | Type | Description |
|---|---|---|
| id | bigserial | PK |
| message_id | varchar(255) | Identifier for the message |
| identification_value | text | Identification value of the organization that is sending the message |
| name | text | Name of the organization that is sending the message |
| payload | text | Payload of the message |
| payload_after_validation | text | Payload after (xsd-)validation |
| response | text | Response to the message (received from myCarenet) |
| valid | boolean | Whether the message is valid or not |
| interface_type | varchar(25) | Type of interface used for the message (e.g. FILE_SYSTEM or REST) |
| status | varchar(25) | Current status of the message (possible states: INVALID (validation against xsd-scheme failed), TO_SEND (ready for sending to myCarenet), SENT (sent to myCarenet), ERROR (something went wrong during sending, e.g. unable to reach myCarenet)) |
| created_on | timestamp | Timestamp of when the message was created |
| input_reference | varchar(255) | Reference for the input message (this is a unique identifier that can be used for debugging/tracing with myCarenet) |
| issuer | text | Issuer of the message |
| postresponse_tack_applies_to | text | Applies-to value for the TACK post-response (received from myCarenet) |
| postresponse_tack_id | text | ID of the TACK post-response (received from myCarenet) |
| postresponse_tack_reference | text | Reference for the TACK post-response (received from myCarenet) |
| postresponse_tack_resultMajor | text | Result major for the TACK post-response (received from myCarenet) |
| postresponse_tack_resultMinor | text | Result minor for the TACK post-response (received from myCarenet) |
| postresponse_tack_resultMessage | text | Result message for the TACK post-response (received from myCarenet) |
| previous_registrationcode | varchar(255) | Previous registration code for the message (obsolete) |
| current_registrationcode | varchar(255) | Current registration code for the message (value will be identical to previous_registrationcode) |
Query 5: Connect to the Nippin database postgresql://<server_ip>:5432/nippin (same user/password) to validate the current state and payload for the nippin message based on the registration code.
select * from nippin_message where current_registrationcode = 'use the value of Query 4 (first query)';This documentation is being updated regularly. We try to provide as correct, complete and clear as possible information on these pages. Nevertheless, if you see anything in the documentation that is not correct, does not match your experience or requires further clarification, please create a request (type : request for information) via our portal (https://sciensano.service-now.com/sp) or send us an e-mail via support.healthdata@sciensano.be to report this documentation issue. Please, do not forget to mention the URL or web address of the page with the documentation issue. We will then adjust the documentation as soon as possible. Thank you!Nippin database
Nippin databaseThe different statuses of a Nippin message in the database
| Status | Description |
| TO_SEND | HD4DP v2 ready to be sent to MyCareNet or SFTP folder |
| ARCHIVED | Messages are set to ARCHIVED, only if the NippinCleanup table has 0 records. These messages will not be processed and won't receive any attention afterwards. |
| INVALID | XML payload is invalid, a ticket can be created at our service portal, including the payload of the invalid message. |
| BUFFERED | not used |
| ERROR | HD4DP v2 was not able to send the message to MyCareNet or SFTP folder |
| SENT | HD4DP v2 was able to send the message to MyCareNet or SFTP folder |
| Column Name | Data Type | Description |
|---|---|---|
| id | bigserial | Primary Key |
| message_id | varchar(255) | Identifier for the message |
| identification_value | text | Identification value of the organization that is sending the message |
| name | text | Name of the organization that is sending the message |
| payload | text | Payload of the message |
| payload_after_validation | text | Payload after (xsd-)validation |
| response | text | Response to the message (received from myCarenet) |
| valid | boolean | Whether the message is valid or not |
| interface_type | varchar(25) | Type of interface used for the message (e.g. FILE_SYSTEM or REST) |
| status | varchar(25) | Current status of the message (possible states : INVALID (validation against xsd-scheme failed), TO_SEND (ready for sending to myCarenet), SENT (sent to myCarenet), ERROR (something went wrong during sending, e.g. unable to reach myCarenet)) |
| created_on | timestamp | Timestamp of when the message was created |
| input_reference | varchar(255) | Reference for the input message (this is a unique identifier that can be used for debugging/tracing with myCarenet) |
| issuer | text | Issuer of the message |
| postresponse_tack_applies_to | text | Applies-to value for the TACK post-response (received from myCarenet) |
| postresponse_tack_id | text | ID of the TACK post-response (received from myCarenet) |
| postresponse_tack_reference | text | Reference for the TACK post-response (received from myCarenet) |
| postresponse_tack_resultMajor | text | Result major for the TACK post-response (received from myCarenet) |
| postresponse_tack_resultMinor | text | Result minor for the TACK post-response (received from myCarenet) |
| postresponse_tack_resultMessage | text | Result message for the TACK post-response (received from myCarenet) |
| previous_registrationcode | varchar(255) | Previous registration code for the message (obsolete) |
| current_registrationcode | varchar(255) | Current registration code for the message (value will be identical to previous_registrationcode) |
| version_tag | varchar(255) | Current application-version |
Granted privileges
| database | user | privileges |
| nippin | dpuser | CONNECT/nippin_message:SELECT/nippin_cleanup:SELECT |
Queries
- Count records grouped by the type and status:
nippin=# select interface_type, status, count(id) from nippin_message group by interface_type,status;
interface_type | status | count
----------------+---------+-------
FILE_SYSTEM | SENT | 117
FILE_SYSTEM | INVALID | 352
FILE_SYSTEM | TO_SEND | 9238
(3 rows)- Get all error and invalid information:
select id, message_id, project_id, dcd_id, payload_after_validation from nippin_message where status in ('INVALID', 'ERROR');- Get previous and current registration code:
select id, message_id, project_id, dcd_id, previous_registrationcode, current_registrationcode from nippin_message;- Get nippin cleanup information:
select * from nippin_cleanup;MyCareNet integration-specific queries
Only for hospitals that are using the Nippin integration in HD4DP v2.
- Count records with status SENT and group them based on the reference ID received from MyCareNet:
select postresponse_tack_result_major, postresponse_tack_reference, count(*) from nippin_message where status = 'SENT' group by status, postresponse_tack_result_major, postresponse_tack_reference;
postresponse_tack_result_major | postresponse_tack_reference | count
-----------------------------------+--------------------------------------+-------
urn:nip:tack:result:major:success | ***** | 2
urn:nip:tack:result:major:success | ***** | 88This documentation is being updated regularly. We try to provide as correct, complete and clear as possible information on these pages. Nevertheless, if you see anything in the documentation that is not correct, does not match your experience or requires further clarification, please create a request (type : request for information) via our portal (https://sciensano.service-now.com/sp) or send us an e-mail via support.healthdata@sciensano.be to report this documentation issue. Please, do not forget to mention the URL or web address of the page with the documentation issue. We will then adjust the documentation as soon as possible. Thank you!HD4DP v2 Online Acceptance Environment
HD4DP v2 Online Acceptance EnvironmentIntroduction
To support development and validation of data transfers using S2S API or CSV upload, a central Online Acceptance Environment is available for the IT services or IT partners of data providers. It is meant to replace the locally installed acceptance environments at the side of the data providers. With the Online Acceptance Environment the three types of data transfer can be tested and validated: data transfer via an API platform, via an SFTP client and via manual input in the study form.
In order to keep the acceptance environment light, it is rebuilt once a week (every Saturday), automatically removing all data that have been entered for testing. The data are stored locally and will not be sent to Healthdata.be infrastructure. The testing is limited to the upload to the HD4DP v2 application.
Application URLs and port
- Front-End Application URL: https://hd4dp.acceptance.healthdata.be
- SFTP host name: sftp.acceptance.healthdata.be and port 2220
- S2S API: https://hd4dp.acceptance.healthdata.be/proxy/api/dcd/payload/submit?organization-id=00&dcd-id=00&version=1
Navigate to and access the Online Acceptance Environment (OACC)
The Online Acceptance Environment can be found on https://hd4dp.acceptance.healthdata.be, which is a publically accessible URL.
On the homepage you are requested to select your organization from the drop-down list in order to proceed.
Log in with the credentials you have received upon request. The username is test@sciensano.be for all users.
Since the list of organizations to choose from is limited, you might not find your organization in it. In that case we advise you to request your credentials through our service portal at https://sciensano.service-now.com/sp via the Request something tab and subsequently the Request for Information box.
When requesting an account for this acceptance environment you will receive 3 different types of credentials:
⦁ credentials to log in to the front end of the online acceptance environment
⦁ credentials to use the API (-> authorization tab in Postman)
⦁ credentials for the SFTP server you use to test the CSV Upload
Once logged in, the layout looks very familiar: to the left you will find the navigation panel with all running projects and projects that have passed the user acceptance testing (UAT) phase. Note that the list of projects featuring in our Online Acceptance Environment is not filtered out for the organization you have selected.
The data transfer methods
As mentioned above, the Online Acceptance Environment enables the testing of the uploads for the following three types of data transfer. They are described in order of preference underneath:
Data transfer via an API platform
The data are extracted directly from the EPD systems and sent to HD4DP v2 local using S2S API before they are sent to healthdata.be. This transfer method requires the use of an API development platform, such as Postman (freely available).
The endpoint (URL) to send your payload to for testing is
https://hd4dp.acceptance.healthdata.be/proxy/api/dcd/payload/submit
This endpoint is to be completed with some parameters, such as the ID of an organization, the ID of a dcd, the version number:
https://hd4dp.acceptance.healthdata.be/proxy/api/dcd/payload/submit?organization-id=6&dcd-id=18&Version=1
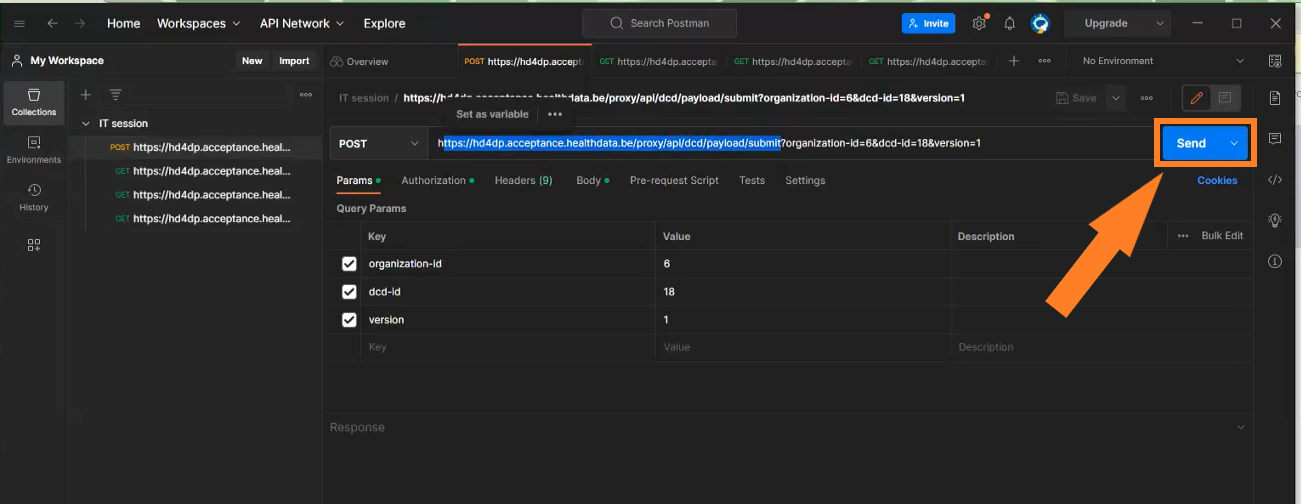
Click on the Send button to post the payload. A succesful submission is indicated with the status message “202 Accepted”. This can also be checked visually in the front end of the Online Acceptance Environment.
The field Data source in the top selection bar indicates whether the data were transferred via S2S API, CSV Upload or manually with HD4DP.
In the production environment records sent through API are sent directly to the healthdata.be infrastructure (status “Submitted” in the Progress field).
Next to posting payloads (POST) you can also retrieve information (GET). Examples of such "calls":
- The call https://hd4dp.acceptance.healthdata.be/proxy/api/organization (see below) will return an organization id:
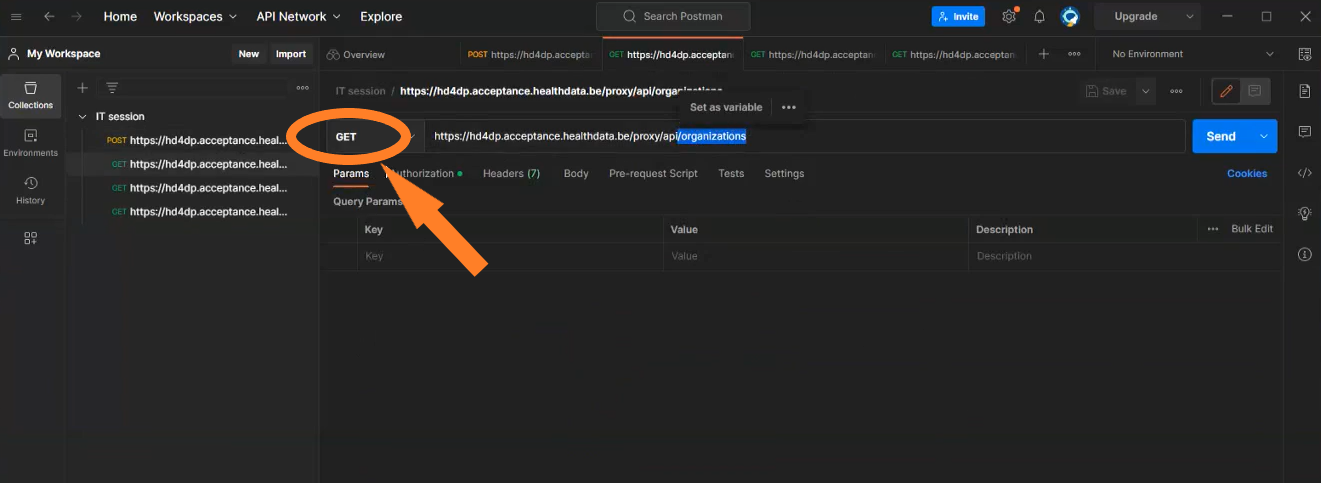
- The call https://hd4dp.acceptance.healthdata.be/proxy/api/dcd/menu/structure?organization-id=6 (see below) will return the menu structure with all projects your organization is registered for.
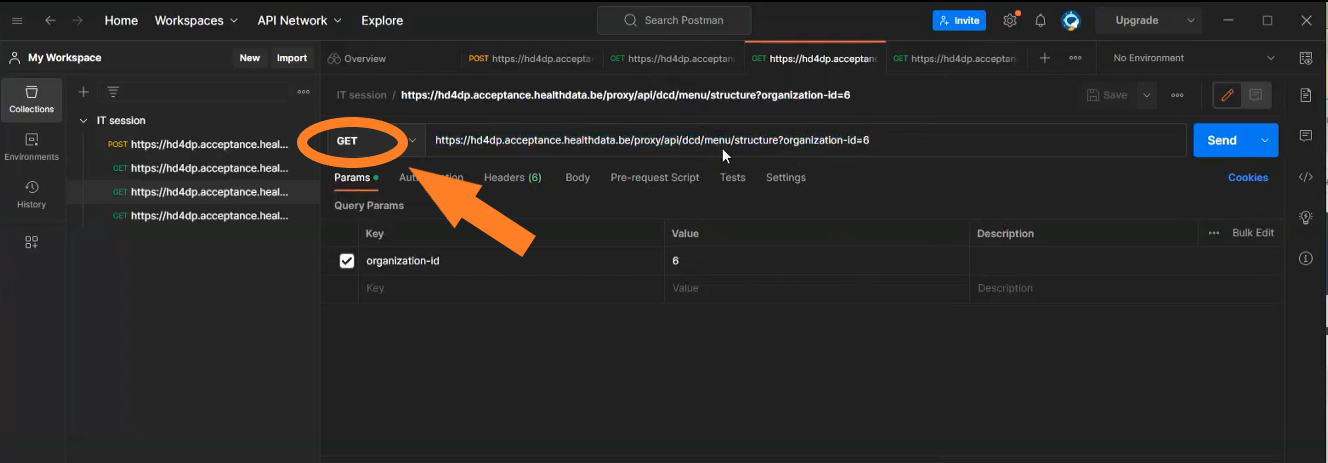
More information about the API data transfer can be found at https://docs.healthdata.be/documentation/hd4dp-v2-health-data-data-providers/hd4dp-v2-s2s-api
Data transfer via an SFTP client
The csv upload is the second type of data transfer. The data are transferred to an SFTP server and subsequently picked up by the healthdata.be system. This transfer method requires the use of an SFTP client, such as WinSCP (freely available).
The Login window will look somehow as follows:
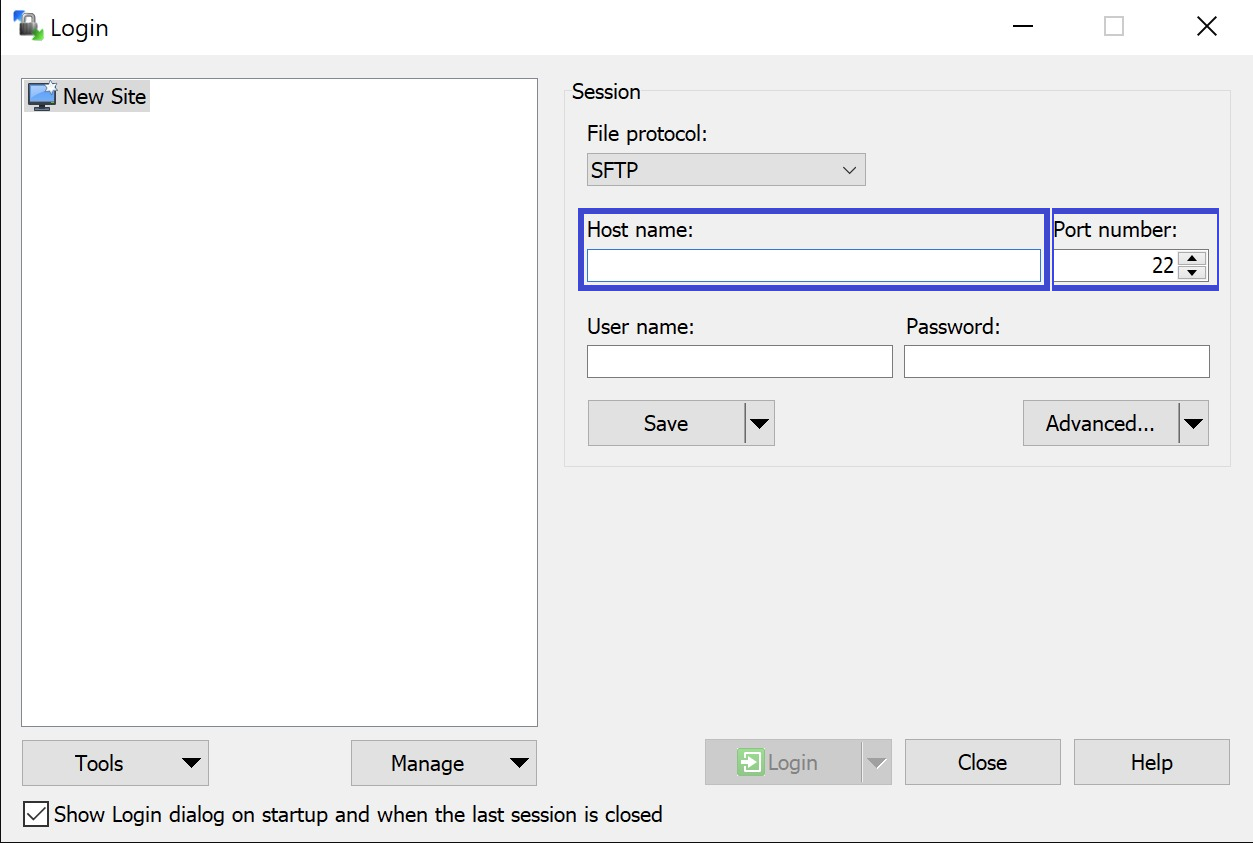
Herein, you enter Host name (sftp.acceptance.healthdata.be) and Port number (2220).
Credentials for the SFTP folder are shared together with the Front-end and API credentials as described in the section “Navigate to and access the Online Acceptance Environment”.
When logging in to WinSCP, you will need to navigate to the correct csv folder : csv/<project>/<dcd>. Here you need to drag and drop the csv you want to upload from the left panel to the right panel. The CSV file will now be picked up by the polling system of the CSV Uploader, which checks for new CSV files every minute.
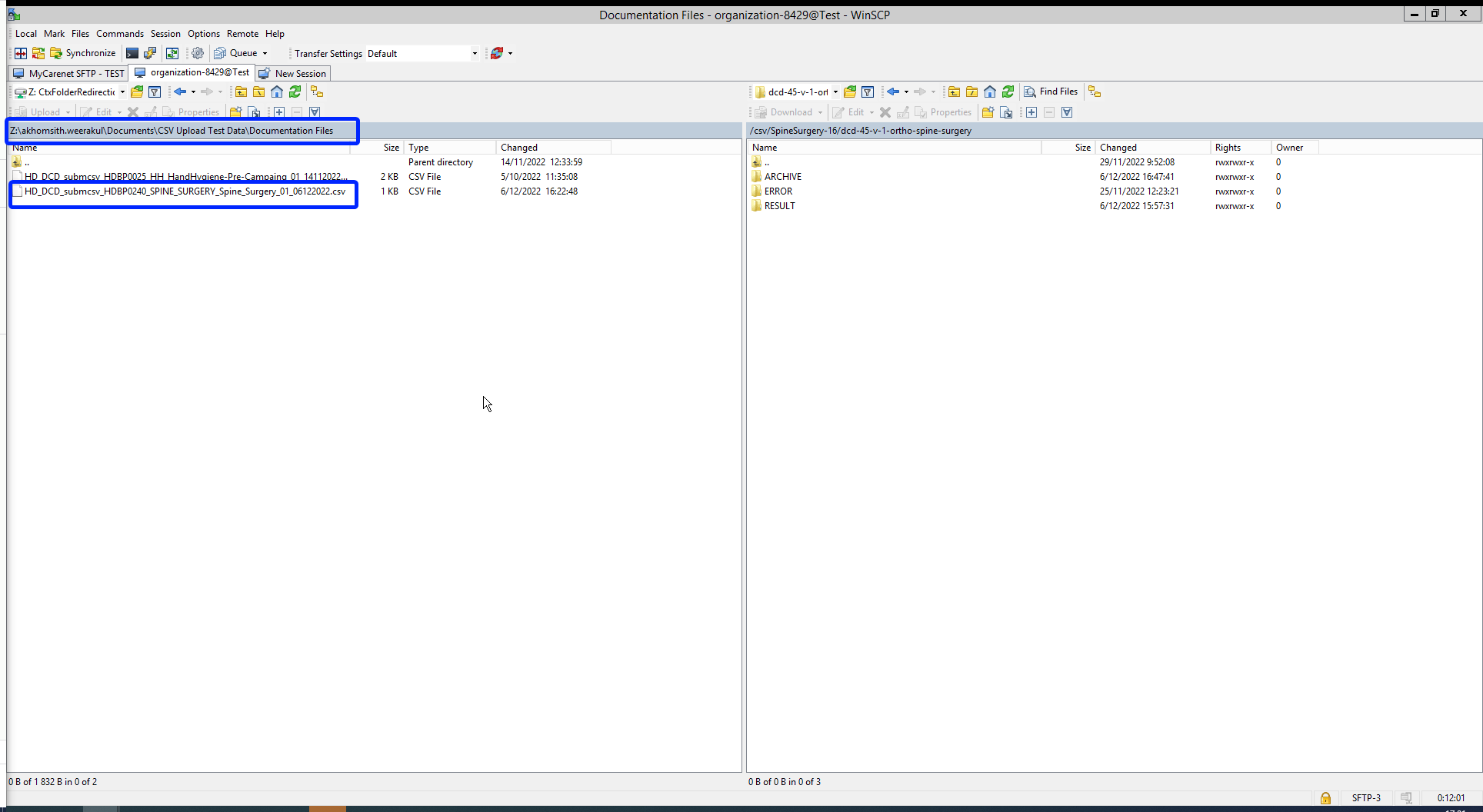
The folders Archive and Result will only be created after the first CSV file has been uploaded for testing.
The Result folder shows a log file containing CSV Upload reports. The status Error Count shows technical errors such as incorrect name, code …
CSV files that were uploaded in Architecure 1 can be reused in Architecture 2. Prerequisite for this is the addition of necessary fields that are typical for Architecture 2, e.g.:
- Author Group (TX_AUTHOR_GR) with the value "Test group"
- Author (TX_AUTHOR) with the value "test@sciensano.be"
- Coauthor (TX_COAUTHOR) with the value "test@sciensano.be"
To further facilitate the process of reusing CSV files a mapping table with old and new CSV names is provided.
Next to adding fields, you can also leave out fields, which is indicated in the log file reports as a warning.
Once again the back-end process can be checked in the study forms on the front-end interface. You want to refresh the window to update to the newest status.
More information about the CSV data transfer can be found at https://docs.healthdata.be/documentation/hd4dp-v2-health-data-data-providers/hd4dp-v2-csv-upload.
Manual input in the study form
The third data transfer method is the form entry, carried out manually. For this you can use a common browser such as Google Chrome. This method can also be used to validate data sent via S2S API or CSV Upload.
This documentation is being updated regularly. We try to provide as correct, complete and clear as possible information on these pages. Nevertheless, if you see anything in the documentation that is not correct, does not match your experience or requires further clarification, please create a request (type : request for information) via our portal (https://sciensano.service-now.com/sp) or send us an e-mail via support.healthdata@sciensano.be to report this documentation issue. Please, do not forget to mention the URL or web address of the page with the documentation issue. We will then adjust the documentation as soon as possible. Thank you!Requesting access
Requesting accessA request for access to the online acceptance environment will be made available in the Entity Access Management tool available via eam.healthdata.be. See also https://docs.healthdata.be/EAM
This feature is foreseen to be added to the EAM portal during the summer of 2023. In the meantime or when the EAM portal should be unavailable, you can use the service portal to request access to the online acceptance environment by creating an incident and clearly mentioning you are an IT Department or IT partner of a data provider requesting access to the online acceptance environment.
More information on how to create an incident in the service portal is available on https://docs.healthdata.be/documentation/hd4dp-v2-health-data-data-providers/how-report-incident
You will receive 3 types of credentials:
- Credentials for the HD4DP2 web form application.
- Credentials for the API data transfer.
- Credentials for the SFTP server to upload CSV files.
Support service for HD4DP v2
Support service for HD4DP v2The Service Desk of healthdata.be (Sciensano) helps users of our applications and services and deals with requests and problems when they arise.
The Service Desk focuses on those services run by our IT Services (HD4DP, HD4RES, healthstat.be,...) and helps you with accounts and passwords. For questions about the content and objective(s) of the projects, we kindly refer to the managing research organizations.
For most efficient processing of your request, we advise you to use our service portal: https://sciensano.service-now.com/sp.
Please find below our support window hours:
How to report an incident
How to report an incidentThe service healthdata.be (Sciensano) handels each report of an incident according to a Standard Operating Procedure (SOP). A public version of this SOP "HD Incident Management Process" is also available on this docs.healthdata.be portal.
To submit an incident related to the projects and applications that are in production, and facilitated or managed by the service healthdata.be of Sciensano, you first need to log in to the HD Service and Support portal.
After the log in step, you will arrive at the main page of the portal.
On the main page, you have to select "Get Help"

A new page with the title "Create Incident" will appear.
You can now document your incident or problem by providing following information:
Please indicate the urgency your problem needs to be resolved according to its business criticality.
Please indicate the type of problem you are experiencing.
When the problem type "Application" is selected, two extra fields appear: "Project name" and "Application".

Please select the appropriate information.
Please describe clearly and briefly (1 sentence) the subject of your problem.

Please describe in detail the problem. Following aspects are important for us to understand and to solve the problem:
- a description of the actions you want to perform but fail to accomplish (Ex: provide us field name, validation rule, button, etc.);
- a description (if possible) of the sequential steps you take to use the healthdata.be service or application you need support for;
- a brief description of the technical problem you are experiencing (e.g. error messages)

We highly recommend to add a screenshot describing the problem (IMPORTANT: do not provide us patient data!).
You can add the screenshot by pressing "Add attachments"
On the right side of the form, the required information elements of the Incident form are listed. When these fields are completed, these field names will disappear in the "required information" box.
Only if all required fields are completed , a form can be submitted., by pressing the green "Submit" button.
If not all required fields were completed, a warning message will appear on top of the form.

Also, the missing required fields will be highlighted in green.

When the incident form was successfully submitted, an overview of your submission will appear in an new screen.
On the right of the screen, you will find the details , including the Incident number.
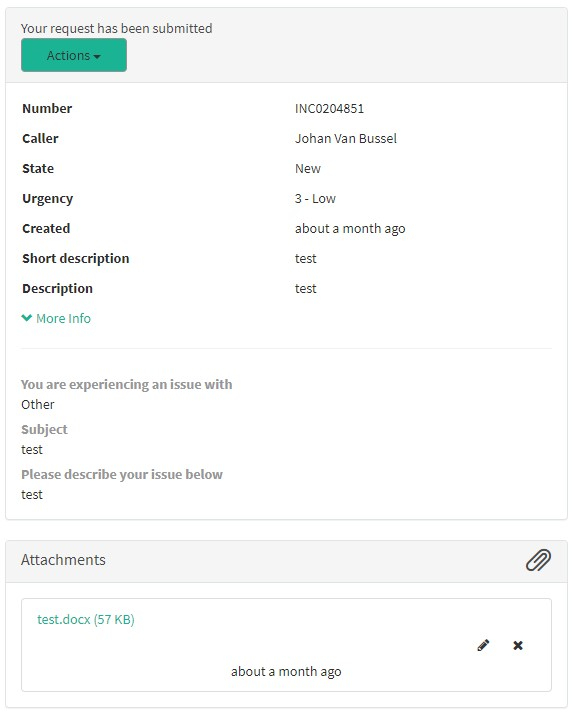
On the left of the screen, you will find a timeline of the handling your incident, starting with your creation.
Release notes
Release notesApril 9th, 2024
Pacemaker fix for nippin was added to production
- Update the status of the invalid messages (INVALID) in the Nippin database to archive (ARCHIVED) as new messages would be created
- For each "invalid" or now archive pacemaker entry in the database, a new record should be created with a TO_SEND or SENT status
February 29th, 2024
Whitelisting URL to avoid registration code issues
- Added URL for whitelisting: https://*.npmjs.org/*
October 18th, 2023
Uniform naming convention
- In the frame of the Architecture 2.5 release a Uniform naming convention has been introduced.
Sending code values instead of code IDs
- In the frame of the Architecture 2.5 release Code values are sent instead of code IDs.
MDM mapping of billingcodes for MyCarenet
- In the frame of the Architecture 2.5 release MDM Mapping of 13 character Billing codes for MyCarenet has been implemented
July 27th, 2023
Whitelisting URL allowing EAM functioning
- Added URL for whitelisting: https://eam-production.servicebus.windows.net/*
Adaptation Port allowing EAM functioning
- TCP 5671, 5672 and 443: eam-production.servicebus.windows.net: HTTPS and AMQP 1.0
Adaptation Port allowing HERA functioning
- TCP 2222: hera-dc.healthdata.be